Long before Darwin and Wallace, people were using selection to change the features of plants and animals. Farmers and breeders allowed only the plants and animals with desirable characteristics to reproduce, causing the evolution of farm stock. This process is called artificial selection because people (instead of nature) select which organisms get to reproduce.
As shown below, farmers have cultivated many crops from wild mustard by artificially selecting for certain attributes.

 Digging Data
Digging Data
Just as humans developed crop plants and domesticated animals from wild ancestors, we also used artificial selection to create distinct varieties and breeds of these species. Domestic dogs evolved from ancient, now-extinct wolf ancestors tens of thousands of years ago[1] – and then, over the last 200 years, humans further selected subsets of dogs, creating Great Danes, Chihuahuas, and the full gamut of more than 450 breeds.

During the process of developing different breeds, the focus was on traits. We didn’t know which genes bestowed, for example, swimming skills or rat-catching ability; we just knew that we wanted dogs that were better at pulling fishing nets (leading to Chesapeake Bay Retrievers and Portuguese Water Dogs) and clearing out pests (leading to the Rat Terrier). In recent decades, scientists have been able to track down some of the genes that we unwittingly selected for in different breeds. Elaine Ostrander, a biologist at the National Institutes of Health, is one of these scientists. She and her team study dogs in order to answer basic questions about genetics that have implications for human health.

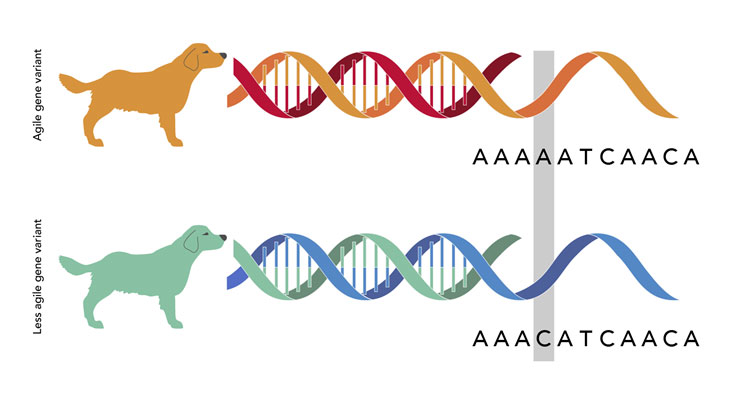
In one study, Elaine and a team that included Jaemin Kim, Falina Williams, Dayna Dreger, Jocelyn Plassais, Brian Davis, and Heidi Parker focused on sport hunting dogs, which are an active group of breeds that perform many different jobs. This group includes spaniels, retrievers, and hunting dogs. The team identified 59 genes that seem to have been the targets of recent selection in these dogs.[2] A particular variant (i.e., an allele) of one of these genes – called ROBO1 – was common in the most agile breeds. These athletic breeds were unusually likely to have an A base at a location in the gene where other breeds were more likely to have a C base. The initial data suggested that ROBO1 was one of the genes that humans unwittingly affected when they artificially selected breeds for different sorts of athletic abilities, but the team wanted more evidence.
Background
Broadly, agility is the ability to move quickly and easily, but for dogs, it can have a more specific meaning. Agility is a sport in which a person directs a dog to run through a complex obstacle course. For a dog to perform well in agility competitions, it must have both the athletic skills to make it through the course quickly and the mental skills to respond nimbly to commands and assess obstacles.

ROBO1 may be related to those mental skills. From studies of humans, we know that ROBO1 is involved with brain development. Different versions of ROBO1 affect how the brain works and learns. For example, having a certain version of ROBO1 seems to contribute to dyslexia, a condition where people have trouble reading.
Hypothesis
The team had identified the A version of ROBO1 based on a relatively small sample of breeds. This led to the hypothesis that the A version of the gene was favored by artificial selection in breeds in which humans desired traits that lead to high agility. If this hypothesis were true across breeds, then we’d expect the A gene version to be more common in more agile breeds – that is, we’d expect to observe a positive relationship between the frequency of the A gene version and breed agility.
Data
To assess agility, the team used records of which breeds were entered in agility competitions and which breeds actually won agility titles. For each breed, they calculated the number of titles won per dog of that breed entered in competition. They then divided breeds into eight groups according to how likely each breed was to win agility titles:
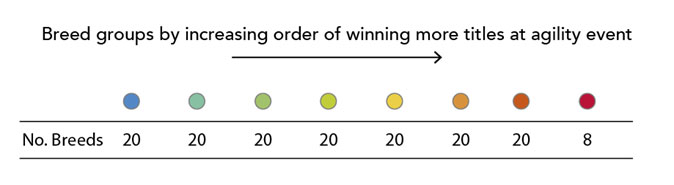
Top performers (such as Border Collies and Pumis, both herding dogs) were in the red group. Breeds that never or very rarely won agility competitions (like the Greyhound and Anatolian Shepherd) were in the blue group.

To figure out how common the A gene version of ROBO1 was in these different groups of dogs, the team turned to a database of dog DNA sequences. They found ROBO1 data for 1243 dogs of breeds in one of the eight breed groups. These were not dogs that had necessarily competed in agility trials themselves; they were just dogs that were members of the same breeds as competitors. For each of the eight breed groups, the team calculated how common the A gene version of ROBO1 was compared to other gene versions. Here is what they found:
On this graph, the x-axis orders the eight breed groups from least likely to win agility titles (far left) to most likely (far right). The y-axis shows how common the A gene version is in that group. So, for example, a frequency of 0.4 means that the A gene version was found in 40% of the sequences from dogs of that group. There is a clear, strong relationship between tendency to win agility titles and frequency of the A gene version: the regression line slopes upwards, and this is significant (p = 0.0038). R2 indicates how well the frequency of different gene versions is explained by the breed group. An R2 of 0 would indicate that breed group cannot predict the frequency of the A gene version at all. An R2 of 1 would indicate that one can precisely predict the frequency of the A gene version based on breed group. In this case, the R2 value of 0.77 indicates that 77% of the variation in the frequency of the A gene version is explained by breed group. These findings further support the hypothesis that the A version of ROBO1 was under strong artificial selection by humans in breeds for which humans desired traits that lead to strong agility. Now researchers can build on this hypothesis to learn exactly how the A version of ROBO1 impacts dogs that carry it, why the A gene version was favored in some breeds but not others, and importantly, how this might be related to athletic performance and other traits in humans.
Stepping into science
When Elaine Ostrander learned about genetics as a sophomore in high school, she knew she’d found her calling. She wanted to be a scientist. But she didn’t have any role models around her: no parent or friends were scientists. So Elaine forged her own path. Throughout high school and college, she worked all sorts of jobs – tutor, waitress, janitor, dishwasher – so she could get the training she needed for her dream career. And she made it. Elaine now leads her own lab, where she helps train the next generation of scientists and builds new knowledge about genetics.
[1]Scientists are still working on untangling exactly how many wolf populations were involved in this process and where and when it occurred.
[2]Identifying these genes was a big part of the study, though not the focus of this Digging Data article. The process involved comparing the genomes of many sport-hunting dogs to the genomes of terriers (breeds selected to hunt vermin). The team looked for areas in the genome where 1) sport-hunting dogs tended to have one sequence and terriers tended to have a different sequence (suggesting that the two groups have evolved genetic differences from one another) and 2) the stretch of DNA sequence common in one of the groups of dogs was unusually long – that is, many dogs in one of the two groups (but not the other) had chromosomes with the exact same long genetic sequence at a particular location. This suggests that somewhere in that sequence is a genetic variant (allele) that was so advantageous that it became common quickly – so quickly that recombination didn’t have time to mix up the sequence near the useful variant. This process is known as a selective sweep. So, in short, the team looked for parts of the dog genome where there was evidence of genetic differences between sport-hunting dogs and terriers and evidence that a selective sweep caused those differences to evolve.
- Youth-produced video from the California Academy of Sciences introducing dog domestication and artificial selection
- TedEd talk on dog domestication
- NOVA program on dog domestication, available through the Teacher Institute for Evolutionary Science
- Middle and high school level lesson on artificial selection in dogs
- High school level lesson on the evolution of domestic dogs from BiteScis
- Student reading with comprehension questions (.docx download)
- Student reading with comprehension and data interpretation questions (.docx download)
- Brief student reading on dog domestication from the Teacher Institute for Evolutionary Science (.doc download)
- Original scientific article from Proceedings of the National Academy of Sciences
For answer keys to the worksheets above, teachers can email UEKeys@berkeley.edu from a school email address and must be verifiably employed as an educator at that school.
The following Data Nugget activity for high school students offers opportunities to analyze data on domestication and selection in hybrids between wild birds and domestic chickens:
Read about:
- Artificial selection in the lab
- Data and error analysis in science: A beginner’s guide (PDF) – includes background information on confidence intervals
- The linear regression test (PDF) – background information
Kim, J., Williams, F. J., Dreger, D. L., Plassais, J., Davis, B. W., Parker, H. G., and Ostrander, E. A. (2018). Genetic selection of athletic success in sport-hunting dogs. Proceedings of the National Academy of Sciences. 115: E7212-E7221.
Reviewed and updated, September 2021

