If Robert Stebbins’ original hypothesis is right, sequence data from Ensatina‘s nuclear DNA should show the same patterns as its mitochondrial DNA, with the most diversity in the north and big genetic differences distinguishing the southernmost lineages. Furthermore, the evolutionary tree based on these DNA sequences should be consistent with a northern origin and southward diversification. Tom followed in Robert’s footsteps, tromping up and down California to catch the salamanders, taking a tiny tissue sample from the tip of the tail, bringing the samples back to his lab, and then extracting the DNA from them.

But Ensatina‘s nuclear DNA was a hard nut to crack. No one had yet made a detailed study of it because of its quirks. Ensatina has a huge genome — roughly six times bigger than ours! — and it’s littered with repetitive DNA — chunks of DNA that probably don’t code for anything and that repeat the same, short genetic sequence over and over again. All that makes their DNA extremely difficult to sequence.
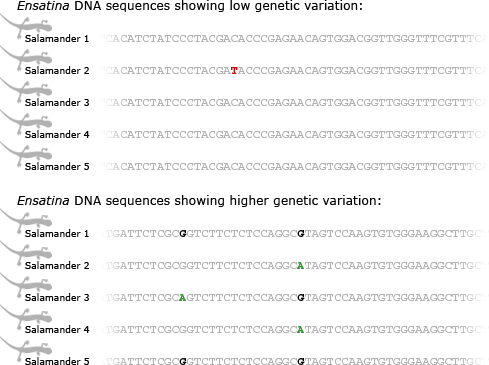
Tom and his colleagues have been working on the project on and off for about four years so far. They have sorted through hundreds and hundreds of short genetic sequences, hunting for ones with enough genetic variation to figure out Ensatina‘s evolutionary history. Different parts of Ensatina‘s genome have different levels of variation — some are remarkably uniform among all lineages, and some vary more from population to population. This is because different parts of the genome evolve at different rates. Since Ensatina has diversified into subspecies relatively recently, the parts of its genome that evolve more slowly haven’t had time to accumulate mutations that would carry information about how the lineages are related to one another. On the other hand, the parts of its genome that evolve more quickly, have accumulated more mutations and now vary among populations. These stretches of DNA can be used to figure out which lineages are most and least closely related.
So far, the team has found ten stretches of DNA that contain useful clues to Ensatina‘s evolutionary history. Tom says that what they are discovering supports the basic hypothesis Robert laid out: “The relationships, for the most part, make sense in terms of being consistent with the morphology and the mitochondrial DNA … but at this point, we need more data.” The extra data will help them figure out exactly how all the subspecies are related to one another and will hopefully paint a more detailed picture of how the salamander populations colonized new areas in their move south.
Get tips for using research profiles, like this one, with your students.