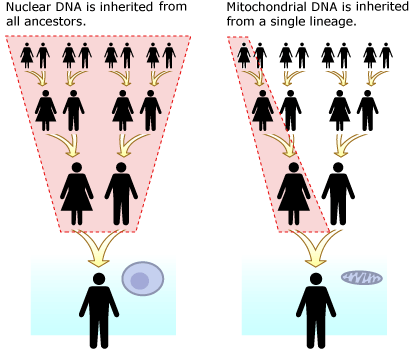
In October 2022, Svante Pääbo was awarded the Nobel Prize in Physiology or Medicine for the work described below.

In the last two months, news outlets have been abuzz with the announcement of what many suggested was a new hominin species. In 2008, a 40,000 year old pinky bone from a child was discovered in a Siberian cave. The bone was not enough to identify the species of its possessor, but since both Neanderthals and humans are known to have lived in the area at the time, scientists assumed it belonged to one of these two species. That all changed in March of this year, when German researchers announced that they’d managed to extract DNA from the fossil — and it didn’t match up to the known genetic sequences of either humans or Neanderthals! In fact, the human and Neanderthal sequences were more similar to each other than either was to the fossil’s sequence! Could this finger bone represent a newly discovered species — nicknamed X-woman? Is the human family tree even bushier than we’d previously thought? Many news outlets touted this research as the first case in which a new species was discovered on the basis of DNA alone. Here, we’ll examine the evidence in more detail and consider what that evidence might — or might not — mean about such claims.
Where's the evolution?
Was X-woman a woman?
Despite the super-heroine nickname, scientists do not know if the new fossil came from a girl or a boy. Mothers pass their mitochondrial DNA on to both their daughters and their sons — so even though the DNA the researchers extracted from the fossil represents a maternal line of descent, we can’t tell the gender of the individual who carried that DNA. If researchers are successful at extracting nuclear DNA from the fossil, they may be able to discover the gender of the child based on the presence or absence of genes from the X and Y chromosomes.
To really understand what this discovery means for our understanding of human evolutionary history, we need to look a little more closely at the evidence. The most important thing to note is that the researchers sequenced the fossil’s mitochondrial DNA — a short loop of DNA housed in the cell’s “powerhouse” organelles, mitochondria. Compared to nuclear DNA, this sort of DNA is easy to get from fossils since each cell originally had many copies of its mitochondrial DNA and only one copy of its main genome. Unlike the rest of our genes, mitochondrial DNA is inherited only from our mothers — and it generally doesn’t undergo recombination. Even though hominin mitochondrial DNA is made up of more than 16,000 individual genetic “letters,” constituting 37 different genes, in terms of inheritance, it behaves more like a single, extremely short gene. Mitochondrial DNA is inherited as a single unit without being broken up. Hence, your mitochondrial DNA represents a single, direct line of descent from your mother’s mother’s mother’s … mother.
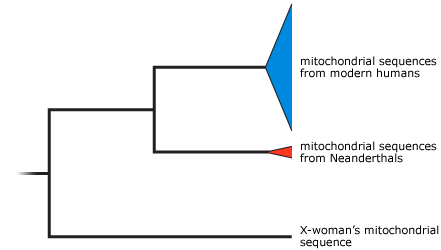
Based on the sequences of mitochondrial DNA extracted from the pinky bone, Neanderthal fossils, and modern humans, the researchers built a phylogenetic tree that shows how all the different sequences are likely related to each other and how recently they descended from a common ancestor. This sort of phylogeny is called a gene tree. It shows the relationships among genes — not necessarily the relationships among species. The gene tree showed that known human and Neanderthal mitochondrial DNA sequences were more closely related to each other than either was to the new sequence from the fossil. Does this mean that the finger bone comes from a new species? Maybe. Maybe not.
If the mitochondrial gene tree turns out to be an accurate indicator of species relationships, it might indeed mean that X-woman is a new species. But it’s also possible that X-woman represents a species distinct from humans and Neanderthals — just not a new species. For example, X-woman might be a member of the species Homo erectus. So far, scientists have not extracted any DNA from known Homo erectus fossils, so we can’t figure this out by looking at X-woman’s genetic sequence. Existing evidence suggests that Homo erectus spread from Africa into Europe and Asia 1.9 million years ago and was unlikely to have lived in Siberia 40,000 years ago — when X-woman lived. However, if we are wrong about where and when Homo erectus (or other known hominin species) lived, X-woman could represent one of these more familiar species.
However, it’s also possible that X-woman’s mitochondrial gene tree doesn’t accurately reflect the evolutionary history of the X-woman species (whatever that species might be). This is because species trees and gene trees don’t always match up. To understand how this can happen, you need to think of species as populations made up of many different individuals:
- At any point in time, there are likely to be many different versions of a gene in the population.

- Most of these gene versions are likely to be closely related to one another.
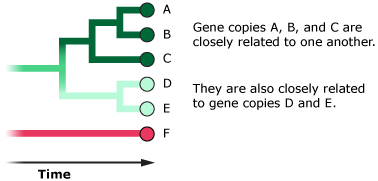
- However, some gene versions may be ancient and may have hung around in the population for a long time.
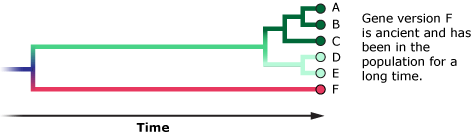
- Sometimes these gene versions are so old that they are actually more closely related to a gene version found in a separate species than they are to other gene versions in the same population.
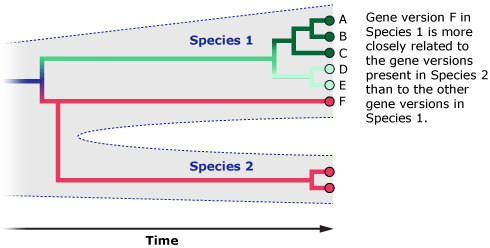
For example, some humans have immune system gene versions that are more closely related to chimpanzee immune system gene versions than they are to other human versions. That’s just because these gene versions are extremely old and have stuck around in both human and chimp populations since our two lineages split from one another six million years ago, not because the humans carrying those genes are more closely related to chimps than to other humans. In this case, the gene tree and species tree don’t match. It’s a bit like if you and your cousin both inherited brown eyes from your grandmother, but your sibling inherited blue eyes from your grandfather. Does this mean that you are more closely related to your cousin than your sibling? Of course not. The genes that control eye color are just a few out of tens of thousands of genes — and if you could look at all of those genes, instead of just one or a few, they would show that you are more closely related to your sibling.
So, what might it mean if the mitochondrial gene tree of X-woman, humans, and Neanderthals doesn’t match the species tree? X-woman might be a human or Neanderthal who happened to carry divergent mitochondrial DNA. This divergent DNA sequence might have been just hanging around in the population since before the human and Neanderthal lineages split — like gene version F in the diagram above. After all, only six Neanderthal DNA sequences are currently available, so we have little idea of how much or how many genetic variants comprised their species. Or perhaps this divergent mitochondrial DNA came from an earlier episode of interbreeding among humans, Neanderthals, or some other hominin. (Some evidence suggests that humans and Neanderthals did interbreed, and some evidence suggests they did not. Scientists are still investigating these possibilities.)
So, with all these possible explanations for X-woman’s unusual mitochondrial DNA, how do we know which is best? We need more evidence. The discovery of new fossils that provide better anatomical information would help us figure out whether or not X-woman belonged to a species we already know about. Unfortunately, that would require a stroke of good luck. So far, scientists have not found any additional fossils, beyond the pinky bone, that could help us. Fortunately, that bone has yet more evidence imbedded in it. The researchers who sequenced its mitochondrial DNA are currently working to extract and sequence nuclear genes from the bone. Since an individual’s nuclear genes come from all its ancestors — not just a single, maternal lineage — this information should give us a much better understanding of how X-woman herself — not just her mitochondrial DNA — is related to humans and Neanderthals. Stay tuned for further updates!
News update, July 2010
As described above, one possible explanation for X-woman’s divergent mitochondrial DNA sequence is that it came from an earlier episode of breeding between different early hominin species. Before this summer, the evidence for mating between humans and Neanderthals was equivocal at best — and the most revealing line of evidence, mitochondrial DNA sequences from several different Neanderthals, suggested that, even if some Neanderthals and humans had interbred, those Neanderthal genes are no longer around. Modern human populations carry no trace of Neanderthal DNA…or so we thought, before we had any nuclear DNA from Neanderthals to study.
In May 2010, an international team of researchers announced that the Neanderthal nuclear genome they’d just sequenced contained a surprise: Neanderthals’ nuclear DNA is more similar to the DNA of modern Europeans and Asians than it is to the DNA of modern Africans! The most likely explanation for this observation is that Neanderthals interbred with the ancestors of Europeans and Asians, possibly when both species lived together in the Middle East — and that Neanderthals didn’t interbreed with the ancestors of modern Africans. The researchers also revealed that many genes from these episodes of interbreeding are still around today. People of European and Asian descent probably share between 1 and 4% of their DNA with Neanderthals! Though this is a significant portion of the genome, it may have resulted from relatively few interbreeding events. After all, at the time these events occurred, the human population was expanding out of Africa and increasing in population size. Genes from just a few interbreeding events may have surfed to relatively high frequencies as the population grew.
So what does this new discovery tell us about X-woman? It still doesn’t indicate whether she represents a new hominin species, but it does highlight the fact that interbreeding among early hominin species happened and that the genetic divisions between species are not always neat. It also emphasizes how informative nuclear DNA can be. The same team that worked on the Neanderthal genome is currently at work on X-woman’s nuclear genome. Stay tuned to find out if her nuclear DNA is as unique as her mitochondrial DNA — and whether this reveals anything more about interspecies breeding between early hominins!
News update, August 2015
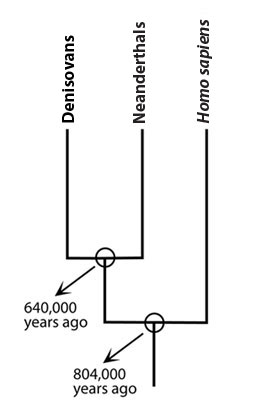 Since we first reported on this story in 2010, X-woman’s lineage has been the subject of a flurry of scientific investigations that have reshaped our view of these early human relatives. First, though, a bit of terminology: the lineage has received a new moniker — X-woman is a Denisovan, a name that comes from the cave in which the fossils were first discovered. Further research in that location uncovered two additional Denisovan fossils: teeth. In addition, scientists have discovered a femur in a cave in Spain that has genetic ties to Denisovans, but there is still uncertainty about to which lineage the original owner of this bone belonged.
Since we first reported on this story in 2010, X-woman’s lineage has been the subject of a flurry of scientific investigations that have reshaped our view of these early human relatives. First, though, a bit of terminology: the lineage has received a new moniker — X-woman is a Denisovan, a name that comes from the cave in which the fossils were first discovered. Further research in that location uncovered two additional Denisovan fossils: teeth. In addition, scientists have discovered a femur in a cave in Spain that has genetic ties to Denisovans, but there is still uncertainty about to which lineage the original owner of this bone belonged.
Scientists have also delved into the evolutionary history of Denisovans by decoding their entire genome — not just the mitochondrial genes. The bulk of the Denisovan genome (i.e., their nuclear DNA) tells a very different story than their mitochondria do. Though Denisovan mitochondrial DNA is equally closely related to Neanderthal and human mitochondria, the rest of their genome is not. It now seems that Denisovans and Neanderthals are more closely related to one another than either is to Homo sapiens.
The other update on Denisovan evolutionary history is that Denisovans interbred with both Neanderthals (about 17% of Denisovan DNA seems to come from Neanderthals) and ancient humans. Denisovans contributed in their own small way to the human lineage through this interbreeding. The highest percentage of Denisovan DNA in modern humans is found in Melanesian populations, which can trace nearly 5% of their genomes back to Denisovans. As results from newly discovered fossils and ancient DNA accumulate, the human branch of the tree of life is getting bushier, and interbreeding seems to be more common than we had previously thought. This might explain why the Denisovan mitochondrial DNA at first led us astray; perhaps X-woman’s mitochondrial genome was introduced to Denisovans through interbreeding with another, as yet unknown hominin lineage.
News update, July 2021
More than a decade ago, the existence of Denisovans was discovered based on DNA extracted from a bone found in a Siberian cave. In the years since, scientists have found more traces of Denisovan DNA — in the Neanderthal and modern human genomes, as well as in remains from that same cave. But we’ve had no direct evidence of Denisovans elsewhere in the world to study…until now. This past year, a team of scientists announced that they’d extracted Denisovan DNA from three separate layers of sediment in a cave on the Tibetan Plateau, suggesting that these ancient human relatives lived there at least on and off for around 50,000 years. The new finding lends further support to the idea that a jawbone from the cave belonged to a Denisovan. The Tibetan cave also contained charcoal from fires, stone tools, and the bones of many other ancient animals, giving us tantalizing clues about the lifestyle of its inhabitants. Because the Denisovans left behind such an obvious genetic legacy in Neanderthals and modern humans, scientists had reasoned that this group must have once been widespread. Now, we have direct evidence of that and an opportunity to learn even more about a lineage that once lived alongside our own.
Primary literature:
- Green, R. E., Krause, J., Briggs, A. W., Maricic, T., Stenzel, U., Kircher, M., . . . Pääbo, S. (2010). A draft sequence of the Neandertal genome. Science. 328: 710-722. Read it »
- Krause, J., Fu, Q., Good, J. M., Viola, B., Shunkov, M. V., Derevianko, A. P., and Pääbo, S. (2010). The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 464: 894-897. Read it »
- Serre, D., Langaney, A., Chech, M., Teschler-Nicola, M., Paunovic, M., Mennecier, P., ... Pääbo, S. (2004). No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biology. 2(3): 0313-0317. Read it »
News articles:
- A review of the discovery from The New York Times
- A blog post exploring possible interpretations of the X-woman DNA from John Hawks' weblog
- Another blog post exploring possible interpretations of the X-woman DNA from The Loom
Understanding Evolution resources:
- Imagine a family composed of a mother, a father, and three biological children: two daughters, and a son. Which of the family members would you expect to have the same mitochondrial DNA sequence? Explain your reasoning.
- List at least three possible explanations for X-woman’s divergent mitochondrial DNA sequence.
- If X-woman is a new species, how would you expect the nuclear DNA from this fossil to compare to the nuclear DNA of Neanderthals and modern humans?
- Review this tutorial on reading evolutionary trees, and examine the phylogeny above. Of modern humans, Neanderthals, and X-woman, which have mitochondrial DNA sequences most closely related to one another? Which have mitochondrial DNA sequences that most recently shared a common ancestor? Explain your reasoning.
- Advanced: Study this figure from the original journal article:
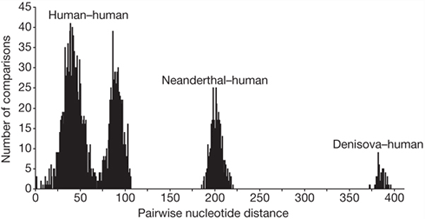
Pairwise nucleotide differences from all pairs of complete mtDNAs from 54 present-day and one Pleistocene modern human, six Neanderthals and the Denisova hominin are shown. Graph reprinted by permission from Macmillan Publishers Ltd: Nature, Krause, J., Fu, Q., Good, J. M., Viola, B., Shunkov, M. V., Derevianko, A. P., and Pääbo, S. (2010). The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 464: 894-897., copyright 2010. To generate this graph, the researchers compared mitochondrial sequences from 55 humans, six Neanderthals, and X-woman (called “Denisova” here). For each comparison, they noted the number of differences between the sequences. The number of genetic differences is shown on the x-axis of this graph. The number of comparisons that had that number of differences is on the y-axis.
- Which pairs of sequences have the most differences? What does this suggest about the relationships among humans, Neanderthals, and X-woman?
- Notice that the human-human comparisons and the Neanderthal-human comparisons form two distinct groups. What does this suggest to you about the relationship between humans and Neanderthals? What might it mean if the two groups of comparisons overlapped by a lot?
- Teach about mitochondrial DNA and human evolution: This interactive and engaging web activity for grades 9-12 compares the number of mutations in mitochondrial genomes to determine ancestry and relatedness of humans and Neanderthals.
- Teach about modern research on hominid and primate evolution: This article for grades 9-12 describes the research of evolutionary biologist Leslea Hlusko. The article reiterates several themes of this news brief, including the interpretation of fossil evidence and the challenge of reconstructing lines of descent from the fossil record.
- Dalton, R. (2010). Fossil finger points to new human species. Nature. 464: 472-473.
- Dalton, R. (2010). Neanderthals may have interbred with humans. Nature. Retrieved April 22, 2010 from Nature.
- Gibbons, A. (2010). Close encounters of the prehistoric kind. Science. 328: 680-684.
- Green, R. E., Krause, J., Briggs, A. W., Maricic, T., Stenzel, U., Kircher, M., . . . Pääbo, S. (2010). A draft sequence of the Neandertal genome. Science. 328: 710-722.
- Krause, J., Fu, Q., Good, J. M., Viola, B., Shunkov, M. V., Derevianko, A. P., and Pääbo, S. (2010). The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 464: 894-897.
- Meyer, M, Fu, Q., Azimu-Petri, A., Glocke, I., Nickel, B., Asuarga, J. Martinez, I., . . . Pääbo, S. (2013). A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 505: 403-406.
- Pennisi, E. (2013). More genomes from Denisova Cave show mixing of early human groups. Science. 340: 799.
- Reich, D., Green, R. E., Kircher, M., Krause, J., Patterson, N., Durand, E. Y.,. . . Pääbo, S. (2010). Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 468: 1053-1060.
- Serre, D., Langaney, A., Chech, M., Teschler-Nicola, M., Paunovic, M., Mennecier, P., . . . Pääbo, S. (2004). No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biology. 2(3): 0313-0317.