Fascination with tiny microbes bearing long, difficult-to-pronounce names is often reserved for biology classrooms — unless of course the bug in question threatens human health. MRSA (methicillin-resistant Staphylococcus aureus) now contributes to more US deaths than does HIV, and as its threat level has risen, so has the attention lavished on it by the media. At this point, almost any move the bug makes is likely to show up in your local paper. Last month saw reporting on studies of hospital screening for MRSA (which came up with conflicting results), stories on MRSA outbreaks (involving both real and false alarms), and media flurries over the finding that humans and their pets can share the infection with one another. Why is this bug so frightening? The answer is an evolutionary one.
Where's the evolution?
MRSA is resistant not only to the antibiotic methicillin, but also to whole other suites of our drugs, making it very difficult to treat and, occasionally, deadly. Modern strains of MRSA did not, however, show up out of the blue. In the early 1940s, when penicillin was first used to treat bacterial infections, penicillin-resistant strains of S. aureus were unknown — but by the 1950s, they were common in hospitals. Methicillin was introduced in 1961 to treat these resistant strains, and within one year, doctors had encountered methicillin-resistant S. aureus. Today, we have strains of MRSA that simultaneously resist a laundry list of different antibiotics, including vancomycin — often considered our last line of antibacterial defense.
How did S. aureus morph from a minor skin infection to a terror? When the media report on MRSA and other drug resistant pathogens, they often say that such pathogens have recently “emerged” — that they’ve “developed” resistance or “learned” to evade our drugs. In fact, it’s more accurate to say that these bugs have evolved resistance. It’s particularly ironic that newspapers might shy away from describing bacterial evolution as such because, when it comes to evolution, bacteria have most of the rest of us beat.
Bacteria are great evolvers for many reasons. For example, their short generation times and large population sizes boost the rate at which they can evolve. In addition, one quirk of bacterial genetics is particularly salient to the evolution of antibiotic resistance: horizontal transfer. Here’s a quick explanation:
- Evolution with vertical transmission. In most familiar organisms, new gene variants arise in a population through random mutation — that is, one individual experiences a genetic mutation and if that mutation ups the individual’s ability to survive and reproduce, it is favored by natural selection. Mutant gene variants are passed from parent to offspring, and advantageous mutations spread through future generations in that way. Over time, additional beneficial mutations that build on the first may occur and begin to spread in the population, allowing more complex traits to evolve as mutations accumulate. This standard picture of evolution is at work in all organisms — whether they are humans that eventually evolve the ability to digest milk or a plant species that adapts to the presence of heavy metals in its environment. The same mechanism also works on bacteria. In fact, biologists have observed the MRSA strain infecting a single patient evolving through random mutation and selection. The patient was being treated with vancomycin, and slowly, over the course of a few months and 35 separate mutations, the bacteria evolved into a vancomycin-resistant MRSA strain.
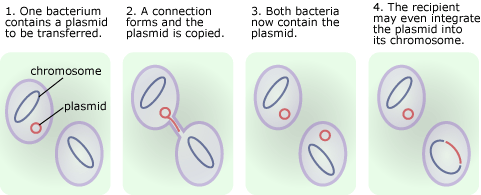
- Evolution with horizontal transfer. So bacteria acquire genetic variation through random mutation, but, unlike humans or oak trees, they also regularly get new gene variants through the process of horizontal transfer — that is, they can pass DNA back and forth to one another directly. For example, bacterial genes can be incorporated into small self-replicating circles of DNA called plasmids, which can be “injected” into other bacteria. The receiving bacterium may even incorporate some of the new DNA from the plasmid into its own genome and pass those genetic sequences on to its descendents. Importantly, bacteria do not have to be closely related to share DNA. Horizontal transfer can occur across even distantly related species — which would be a bit like you picking up the family pet and winding up with a few cat genes in your genome. In terms of evolution, this means that bacteria do not have to rely on random mutation to produce a beneficial gene variant. One species might pick up an advantageous gene from another species, and the process of natural selection could begin to act right away, spreading the new variant through future generations.
Horizontal transfer has important implications for the evolution of antibiotic resistance in bacteria.
- Horizontal transfer can speed up the evolution of antibiotic resistance. Bacteria don’t necessarily have to wait for the right random mutations to come along to, say, adapt to penicillin. If another species or strain has already gone through the process of accumulating the mutations to resist penicillin, the appropriate resistance genes are probably already floating around in bacterial populations. This means that a population that encounters penicillin for the first time may already include a few individuals that carry highly-evolved resistance genes, allowing the strain to evolve quickly to evade the antibiotic.
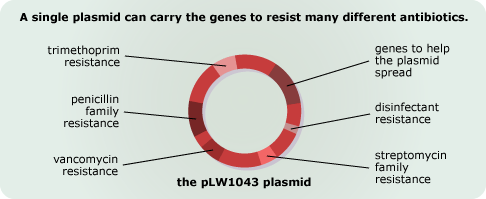
- Horizontal transfer aids the evolution of multiply-resistant strains. Plasmids can carry several genes, and so, separate genes to resist several different antibiotics can make their way onto the same plasmid. For example, in 2003 biologists discovered a single MRSA plasmid that conferred resistance to the penicillin and streptomycin families of antibiotics, as well as two other antibiotics and a disinfectant commonly found in wet wipes! Since the genes are physically attached together, selecting for one of those resistance genes lets the others hitchhike to high frequency. So exposing a bacterial population to say, streptomycin, may also unintentionally favor the evolution of a strain that resists the other antibiotics as well.
Horizontal transfer has played a key role in the evolution of the MRSA strains that currently threaten those in hospitals and, more occasionally, the broader community. S. aureus was treatable with methicillin until a few different individuals obtained an early version of the resistance gene from distantly related species through horizontal transfer. In environments where methicillin resistance was a boon, the gene spread, and regular, old S. aureus evolved into MRSA. The MRSA strains that happened to have the right traits to do well in hospitals thrived in that environment — where they, coincidentally, had the perfect opportunity to pick up additional resistance genes from other hospital-dwelling bacterial species that had already been exposed to our high-end antibiotics. In fact, scientists even successfully (but unhappily) predicted the course of this evolution. In 1992, researchers working in a lab found that Enterococcus faecalis (which inhabits our intestinal tract) were able to transfer genes for vancomycin resistance to S. aureus. They predicted that we would eventually see the same thing happen in a hospital since both E. faecalis and S. aureus are common there and face selective pressure to survive an onslaught from vancomycin. In 2002, it happened. Biologists analyzed a vancomycin-resistant S. aureus strain infecting a Michigan dialysis patient and found that it was the product of horizontal transfer: the bacteria had picked up their resistance genes from E. faecalis.
Faced with the super-evolutionary abilities of bacteria, the situation might seem hopeless — and indeed it is dire. However, there are some simple precautions we can each put into action. Taking antibiotics only for serious bacterial infections and completing your full course of antibiotics as instructed by your doctor will help forestall the spread of antibiotic resistant bacteria. Reducing or eliminating the preventative use of antibiotics on livestock and crops can also help curb the further evolution of resistant strains. And, of course, devoting more research funds to the development of new antibiotics may help tip future skirmishes in this ongoing battle in our favor. But what can we do today about multidrug resistant bacterial strains like MRSA? Should we worry about walking outdoors or shaking hands with a stranger? No. Outside of a hospital, most of us are very unlikely to encounter untreatable strains of MRSA, and something as simple as washing your hands can go a long way towards keeping you healthy — and off the antibiotics that would contribute to the evolution of even more resistant bacterial strains.
Primary literature:
- Chambers, H. F. (2001). The changing epidemiology of Staphylococcus aureus? Emerging Infectious Diseases 7(2):178-182.
- Enright, M. C., Robinson, D. A., Randle, G., Feil, E. J., Grundmann, H., and Spratt, B. G. (2002). The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proceedings of the National Academy of Sciences 99(11):7687-7692. Read it »
- Ferber, D. (2003). Triple-threat microbe gained powers from another bug. Science 302(5650):1488.
- Herold, B. C., Immergluck, L. C., Maranan, M. C., Lauderdale, D. S., Gaskin, R. E., Boyle-Vavra, S., Leitch, C.D, and Daum, R. S. (1998). Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. Journal of the American Medical Association 279(8):593-598.
- Hiramatsu, K., Cui, L., Kuroda, M., and Ito, T. (2001). The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends in Microbiology 9(10):486-493.
- Mwangi, M. M., Wu, S. W., Zhou, Y., Sieradzki, K., de Lencastre, H., Richardson, P., Bruce, D., Rubin, E., Myers, E., Siggia, E. D., and Tomasz, A. (2007). Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proceedings of the National Academy of Sciences 104(22):9451-9456. Read it »
- Weigel, L. M., Clewell, D. B., Gill, S. R., Clark, N. C., McDougal, L. K., Flannagan, S. E., Kolonay, J. F., Shetty, J., Killgore, G. E., and Tenover, F. C. (2003). Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302(5650):1569-1571. Read it »
News articles:
- A news article on recent efforts to screen hospital patients for MRSA from Scientific American
- An overview of the evolution of drug resistance from the New York Times
Understanding Evolution resources:
- What is the key difference between vertical transmission and horizontal transfer?
- Explain how mutation and horizontal transfer affect the genetic variation in a bacterial population.
- Review the process of natural selection. Explain why genetic variation is so important to this process.
- Describe a situation in which treating a bacterial infection with tetracycline would favor the evolution of vancomycin-resistant bacteria.
- Imagine that a friend tells you that penicillin didn’t work on his pneumonia because he’d developed resistance to the drug. Write a short paragraph explaining whether this explanation is correct or incorrect and why.
- Imagine that a friend gets a stuffy nose and a sore throat and decides to take some leftover penicillin in the medicine cabinet to treat the illness. Write a short paragraph explaining whether this is likely a good idea or a bad idea. Give at least three reasons that support your point of view and be sure that that your answer deals with bacterial evolution.
- Teach about the evolution of antibiotic resistance (select the Marc Lipsitch video): In this video for AP biology students, Professor Marc Lipsitch explains how bacteria evolve resistance and why this poses such a threat.
- Teach about natural selection and antibiotic resistance: In this activity for grades 9-12, students learn why evolution is at the heart of a world health threat by investigating the increasing problem of antibiotic resistance in such menacing diseases as tuberculosis.
- Teach about computer modeling and antibiotic resistance: This article for grades 9-12 examines how the scientist Carl Bergstrom uses computer modeling to understand and control the evolution of antibiotic resistant bacteria in hospitals.
- Chambers, H. F. (2001). The changing epidemiology of Staphylococcus aureus? Emerging Infectious Diseases 7(2):178-182.
- Enright, M. C., Robinson, D. A., Randle, G., Feil, E. J., Grundmann, H., and Spratt, B. G. (2002). The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proceedings of the National Academy of Sciences 99(11):7687-7692.
- Ferber, D. (2003). Triple-threat microbe gained powers from another bug. Science 302(5650):1488.
- Herold, B. C., Immergluck, L. C., Maranan, M. C., Lauderdale, D. S., Gaskin, R. E., Boyle-Vavra, S., Leitch, C.D, and Daum, R. S. (1998). Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. Journal of the American Medical Association 279(8):593-598.
- Hiramatsu, K., Cui, L., Kuroda, M., and Ito, T. (2001). The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends in Microbiology 9(10):486-493.
- Mwangi, M. M., Wu, S. W., Zhou, Y., Sieradzki, K., de Lencastre, H., Richardson, P., Bruce, D., Rubin, E., Myers, E., Siggia, E. D., and Tomasz, A. (2007). Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proceedings of the National Academy of Sciences 104(22):9451-9456.
- Sack, K. (2007, October 16). Deadly bacteria found to be more common. The New York Times. Retrieved March 20, 2008 from The New York Times
- Weigel, L. M., Clewell, D. B., Gill, S. R., Clark, N. C., McDougal, L. K., Flannagan, S. E., Kolonay, J. F., Shetty, J., Killgore, G. E., and Tenover, F. C. (2003). Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302(5650):1569-1571.