Though we often think of evolution as occurring at a snail’s pace, one fish species is highlighting just how quickly evolution occurs — in the right circumstances. Between 1947 and 1976, General Electric released more than a million pounds of PCBs into the Hudson River. PCBs can kill fish and seabirds and have been linked to cancer and other serious health problems in humans. PCBs were banned in 1979, but the toxins have remained at high levels in the Hudson because they settle into the sediments on the bottom of the river and don’t break down. Now, scientists have discovered that, over the past 60 years, one bottom-feeding fish species, the Atlantic tomcod, has evolved resistance to PCBs. Though this evolutionary shift is good news for tomcod, it may put the rest of the food web, which depends on this species, in jeopardy.
Where's the evolution?
Hudson River tomcod evolved resistance to PCBs in the same way that a population of bacteria may evolve resistance to antibiotics over the course of a few hours and in the same way that a mammal species might evolve a camouflaged coat over the course of millennia: natural selection. Though the timescales involved are different, the basic process works the same way. Mutation generates random genetic variation (e.g., an individual with a mutation that happens to protect the organism from PCBs) in a population. When the population experiences a challenge (e.g., exposure to high levels of PCBs), individuals carrying genes that allow them to better survive and reproduce in that situation are favored and leave behind more offspring, which also carry those genes. Over many generations, advantageous genes (e.g., genes that code for resistance to PCBs) become more and more common in the population.
When most fish embryos are exposed to PCBs, the fish develop small hearts that don’t beat normally, and these heart defects seem to cause a host of other complications, which can lead to death. However, Hudson River tomcod don’t have these problems — despite the sky high PCB levels to which they are exposed. Based on these observations and other studies, scientists hypothesized that the tomcod has experienced natural selection favoring PCB resistance, but they didn’t know exactly what sort of adaptation would allow the tomcod to survive with such high PCB levels — until now.
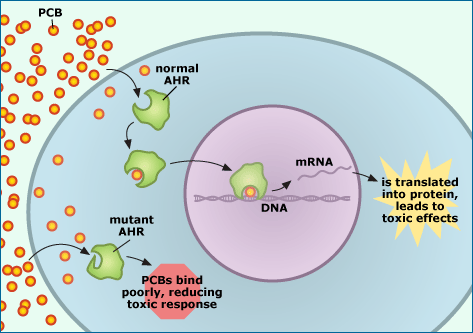
Last month, scientists announced that they’d uncovered the genetic basis of this adaptation. All vertebrates have genes that code for receptors called AHRs (aryl hydrocarbon receptors). These receptor proteins are present in the cytoplasm of cells. When PCBs diffuse into cells, they bind to the AHR protein and are carried into the nucleus of the cell. In the nucleus, the receptor and toxin (along with another molecule they picked up along the way) attach to the cell’s DNA and turn on genes that shouldn’t be activated. This pathway (and perhaps others) seems to be responsible for PCBs’ toxic effects. However, this pathway is blocked in Hudson River tomcod because of a mutation in the gene that codes for AHR. The receptor produced by the mutant gene version doesn’t bind to PCBs easily and so doesn’t travel to the nucleus and activate genes at the wrong times. And this seems to stop the chain reaction that leads to PCBs’ toxic effects.
The mutation responsible is a small one — a six base pair deletion that produces a protein just two amino acids shy of the normal receptor (which is made up of about 1000 amino acids in total) — but the effect is enormous. Tomcod from other rivers are 100 times more sensitive to PCBs than are tomcod from the Hudson! It’s easy to see both how random mutation could cause a six base pair deletion — and how big an advantage fish with just the right six base pair deletion would have in a PCB-contaminated environment.
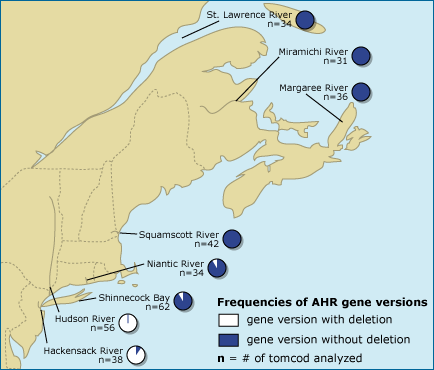
The scientists found evidence of this advantage when they compared the genetic makeup of the Hudson River population to other populations in less polluted rivers. Almost all the fish sampled from the Hudson and the nearby Hackensack River carried the six base pair deletion, but the mutation was extremely rare in other populations. This is exactly what we’d expect to observe if selection favoring the mutation were strong in the Hudson and if the mutation were not advantageous in less polluted waters. Since the Hudson and the Hackensack Rivers share an estuary, these two tomcod populations are linked and can pass genes back and forth — so it is no surprise that the Hackensack population also includes many individuals with the mutation.
While this mutation allows the Hudson River tomcod to thrive in toxic waters, its effect on other organisms in the ecosystem is likely not so positive. Tomcod that carry the mutation can withstand high levels of PCBs, which are stored in the fishes’ fat. When PCB-laden tomcod are eaten by other fish species and those fish are eaten by birds and other organisms, PCBs are passed up the food chain — to species that don’t have the genetic defense that the tomcod do. Though it is tempting to think that those other species will simply evolve resistance to PCBs as well, evolution doesn’t work this way. Natural selection does not automatically provide organisms with the traits they “need” to survive. Some species (like the tomcod) may be lucky enough to have the right sorts of genetic variation and short enough generation times to evolve in response to rapid environmental changes, like PCB contamination — but many others do not. Swift evolution may have saved the tomcod, but it’s no silver bullet. A surer bet is the PCB cleanup project that has already begun and which involves dredging the Hudson to remove these persistent toxins once and for all.
Primary literature:
- Wirgin, I., Roy, N. K., Loftus, M., Chambers, R. C., Franks, D. G., and Hahn, M. E. (2011). Mechanistic basis of resistance to PCBs in Atlantic tomcod from the Hudson River. Sciencexpress. Read it »
News articles:
- A popular article on the research from Wired
- A radio program about the study from NPR.
Understanding Evolution resources:
- A review of the process of natural selection
- Explanations of common misconceptions about natural selection
- A review of DNA and mutations
Background information from Understanding Global Change:
- Review the process of natural selection. Use the four steps described on that page to explain how PCB resistance became so common among tomcod in the Hudson.
- In this blog post, the author describes the tomcod as “a quick learner” because of the population’s adaptation to PCBs. Is “learning” an accurate way to describe the change in the Hudson River tomcod population? Why or why not?
- The data shown on the map above support the hypothesis that natural selection for PCB resistance has occurred among tomcod in the Hudson. What sort of evidence regarding gene frequencies in different populations would argue against this hypothesis (i.e., imagine what the scientists would have observed in this study if natural selection on the AHR gene had not occurred among Hudson River tomcod)?
- Mutations are random in the sense that which mutations occur cannot be predicted based on which traits would be advantageous for the organism. Explain what this means with respect the PCB-resistance mutation in tomcod.
- Advanced: Review the idea of random mutation, paying special attention to the Lederberg experiment. Is there any evidence presented on the map above that supports the idea that the mutation for PCB resistance occurred randomly? If yes, explain what the evidence is and how it supports the idea. If no, explain what evidence would support the idea of random mutations.
- Advanced: The map in the article above shows that the PCB-resistance mutation is present at low levels in populations outside the Hudson and Hackensack Rivers. Formulate a hypothesis that includes the idea of a one-time random mutation and that explains the current distribution of this allele.
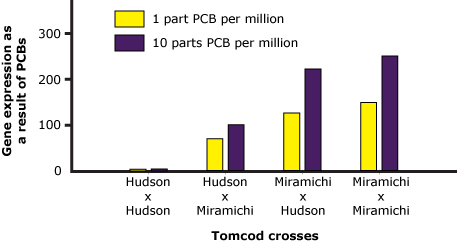
- Advanced: Examine the graph below. It shows how much the offspring of different tomcod crosses (among fish from the contaminated Hudson and fish from the uncontaminated Miramichi) express a particular gene in response to exposure to different levels of PCBs. This is one of the genes that is activated inappropriately by the non-mutant version of the AHR receptor in the presence of PCBs. You can assume that its activation is negatively correlated with fitness in PCB-contaminated waters. Based on these data, would you describe the mutant AHR allele as dominant, recessive, codominant, overdominant, or underdominant? Explain your reasoning.
- Teach about natural selection: In this classroom activity for grades 9-16, students simulate breeding bunnies to show the impact that genetics can have on the evolution of a population of organisms.
- Teach about rapid evolution in other fish populations: This research profile for grades 9-16 follows scientist David O. Conover as he investigates the impact of our fishing practices on fish evolution and discovers what happened to the big ones that got away.
- Teach about rapid evolution resulting from human-caused changes in the environment: This news brief for grades 9-12 describes how global warming has already begun to affect the evolution of several species on Earth.
- Hansson, M. C., Wittzell, H., Persson, K., and von Schantz, T. (2004). Unprecedented genomic diversity of AhR1 and AhR2 genes in Atlantic salmon (Salmo salar L.). Aquatic Toxicology. 68: 219-232.
- Revkin, A. C. (May 15, 2009). Dredging of pollutants begins in Hudson. The New York Times. Retrieved March 8, 2011 from The New York Times.
- Wirgin, I., Roy, N. K., Loftus, M., Chambers, R. C., Franks, D. G., and Hahn, M. E. (2011). Mechanistic basis of resistance to PCBs in Atlantic tomcod from the Hudson River. Sciencexpress. Epublication ahead of print, February 17, 2011.