Now we’ll go through a simple example based on the steps just described.
- Choose the taxa. You decide to study the major clades of vertebrates shown in the leftmost column of the table below. (Note that many vertebrate lineages are excluded from this example for the sake of simplicity.)
- Determine the characters. After studying the vertebrates, you select a set of traits, which seem to be homologies, and build the following data table to record your observations. (Note that many relevant vertebrate characters are excluded from this example for the sake of simplicity.)
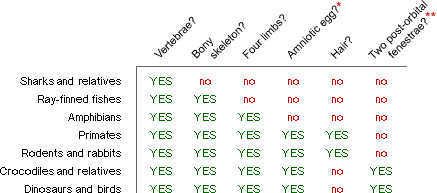
- Determine the polarity of characters. From studying fossils and outgroups closely related to the vertebrate clade, you hypothesize that the ancestor of vertebrates had none of these features.
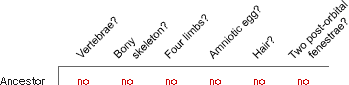
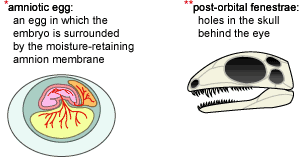
- Group taxa by synapomorphies. Since we have a good idea of what the ancestral characters are (see above), this is not so hard. We might start out by examining the egg character. We focus in on the group of lineages that share the synapomorphic form of this character, an amniotic egg (A, below), and hypothesize that they form a clade (B):
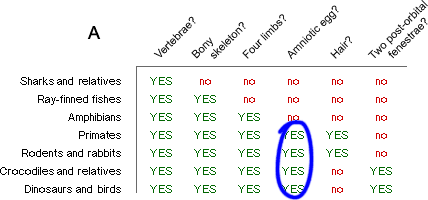
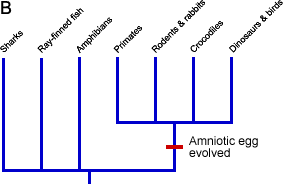 We go through the whole table like this, grouping clades according to synapomorphies (C):
We go through the whole table like this, grouping clades according to synapomorphies (C):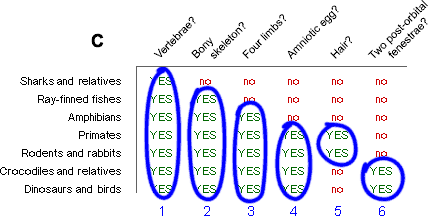
- Work out conflicts that arise. There are no conflicts here. Every group is a subset of another group (see C above).
- Build your tree. Based on the groupings above, you produce this tree:
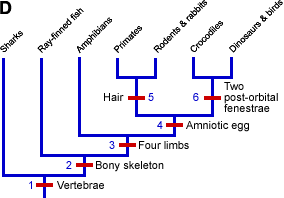
- Voila! You have made a phylogeny.
Of course, this was just an example of the tree-building process. Phylogenetic trees are generally based on many more characters and often involve more lineages. For example, biologists reconstructing relationships between 499 lineages of seed plants began with more than 1400 molecular characters!
