The two possibilities (low replication or no replication) generate very different expectations about what we should observe in a patient’s viral population over the course of the experiment:
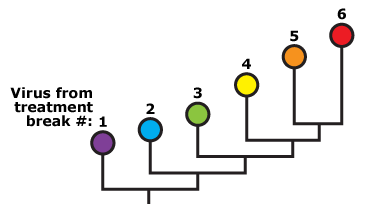
- If HIV replicates at low levels during treatment and rebounds by revving up replication, then … we should observe slow evolution of the virus away from the ancestral viral sequence over time. The virus sampled during the first treatment break should be closely related (and similar) to the ancestral sequence, while virus sampled during later treatment breaks should be more and more distantly related to the ancestral sequence. Here, we’ve shown what this might look like on an idealized phylogeny. Branch length is proportional to the amount of genetic differentiation, and the time of sample collection is represented by different colors.
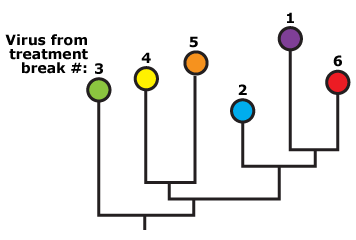
- If HIV does not replicate during treatment and rebounds from the random reactivation of latently infected cells, then … we should not observe any particular pattern related to time of sampling. During some treatment breaks, a viral clone distantly related to the ancestral sequence might be reactivated, and during other treatment breaks, a viral clone closely related to the ancestral sequence might be reactivated. This hypothetical phylogeny shows no pattern related to time of sampling.
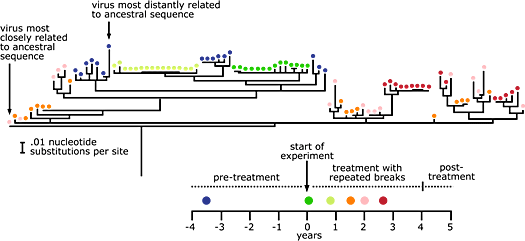
Satish used HIV env sequences to build phylogenies for viruses sampled from nine people over the course of the experiment. All of the phylogenies ended up looking much like that of the patient shown below. Although more viruses are sampled at each time point than in the idealized phylogenies shown above, the big message about viral evolution is pretty clear: there’s no time-related pattern in the data. This random scattering of colors is exactly what we would expect to see if cells that were infected during HIV’s initial infection stage were arbitrarily reactivated during treatment breaks — as Satish puts it, like “random firecrackers going off.”