Most of us get in line at the ice cream shop or tear into a piece of chocolate cake without giving much thought to why we like what we do. Humans appreciate a wide variety of tastes because of our omnivorous evolutionary history and the genes we carry that allow us to sense sweet, salty, sour, bitter, and umami (i.e., savory) flavors. But the same isn’t true of all animals. Most cats, for example, dubiously sniff at sweets. This is because, over the course of its evolutionary history, the feline lineage lost a functional gene to detect sweet flavors. Birds also lack this gene and, usually, the sweet tooth that comes with it — but there are a few notable exceptions. Hummingbirds, for example, make a proverbial beeline for honey-sweet liquids. Why are hummingbirds sugar junkies, while robins stick with worms? New research reveals at a genetic level the evolutionary changes that account for such diverse tastes.
Where's the evolution?
To understand the evolution of taste, you first need to know a little about the biology of flavor. In vertebrates, bitter, sweet, and umami flavors are detected by particular proteins on our taste buds. The bitter taste detector (called a receptor) is composed of a single protein, and the sweet and umami receptors are each made of two proteins linked together. Each of these individual proteins is produced by a different gene. Mutations that change the DNA sequence of those genes can also change which foods our different taste receptors respond to. This is important because taste allows animals to try to get more of a particularly calorie-dense or nutritious food and to avoid compounds that might be harmful or toxic. Without receptors for a particular flavor, animals cannot be motivated to eat or avoid substances with that flavor.
Vertebrates have many different genes that help us sense bitter tastes. Humans, for example, produce 25 separate proteins (encoded by 25 separate genes) that are sensitive to bitter flavors, while rats produce 37 and dogs 16. These diverse bitter-sensing proteins arose through the process of duplication and divergence. Mutations copied an ancestral bitter-sensing gene, giving us multiple versions of this gene in our genomes. Then additional mutations in later generations changed the sequences of those copies in different ways, leading to multiple genes sensitive to different bitter molecules. During this process, the vertebrate lineage was diversifying with mammals, birds, turtles, etc. splitting off at different points and winding up with different numbers of bitter-sensing genes. Scientists aren’t sure why natural selection kept so many of these genes around and functioning, but some think that having many of these genes may allow us to sense and avoid a variety of different toxic compounds in our environments.
In contrast to bitter receptors, sweet- and umami-sensing proteins are remarkably conserved — that is, their number and sequence haven’t changed much over evolutionary time. Most vertebrates have three genes for sensing sweet and savory flavors — T1R1, T1R2, and T1R3. When T1R3 pairs with T1R1, the resulting receptor senses umami; when T1R3 hooks up with T1R2, the structure senses sweet. These three genes are the result of an ancient case of duplication and divergence that occurred more than 400 million years ago, before any vertebrate set foot on dry land. You can see this on the evolutionary tree below. Most phylogenetic trees aim to show the relationships among lineages of organisms, but gene trees like this one show the relationships among genes. The tree shows that two gene duplication events happened before the ray-finned fish and other vertebrates diverged and that the three gene versions were then inherited by most vertebrates.
Since those early duplication events, the sweet- and umami-sensing genes have been faithfully passed down from generation to generation with a few exceptions. For example, in felines, a mutation turned the T1R2 gene off — and with it, cats’ appetite for sweets. The remnants of T1R2 can still be found in feline DNA, but the gene does not function at all. Nonfunctional versions of ancient genes that remain in the genome are known as pseudogenes. In birds, evolution seems to have taken a slightly different course to the same destination. Birds lack any remnant of T1R2 — even a non-functioning one — and so, as a general rule, don’t favor sweet foods, preferring seeds, plants, insects, worms, and other small animals. The mutation that caused the wholesale deletion of this gene occurred in an ancestor of birds that likely lived while massive dinosaurs still walked the earth and was then passed down to all living birds.
But that’s not the end of the story for birds. While most birds can’t taste sweet foods, some can. Hummingbirds far prefer sugary water to plain water, seem to hate artificial sweeteners like aspartame, and yet have no copy of T1R2. How do they sense these different tastes? New research suggests that sometime between 42 and 72 million years ago, when the hummingbird lineage arose from insect-eating ancestors, the hummingbirds’ copies of T1R1 and T1R3 — genes that are typically responsible for umami sensitivity — experienced a set of mutations. The new mutations made the proteins that the genes produced more sensitive to sugars and less sensitive to umami flavors. As the hummingbirds shifted to a nectar-based diet, such mutations were favored by natural selection and the genes were further refined for sweet sensitivity. Today, the evolutionarily repurposed T1R1 and T1R3 genes don’t function exactly as humans’ sweet-sensing genes do (we taste aspartame as sweet, but hummingbirds don’t) but perform the same basic job — orienting their bearers towards the dense calories available in sugary foods. Biologists have yet to figure out the evolutionary explanation for other birds that like sugar — such as the fruit-loving toucan — but it may be that changes in the T1R1 and T1R3 genes are also responsible.
Exploring the evolution of taste perception in vertebrates illuminates the many not-so-mysterious ways in which evolution works. As genetic technologies improve, we are uncovering a detailed picture of the various mechanisms by which genes are gained and change functions, as well as how they are lost. Within this single example, we’ve explored multiple cases of duplication and divergence, genes that were deleted wholesale, genes that were turned off but stuck around in the genome accumulating mutations, genes that were maintained nearly as they started by natural selection, and genes that were nudged towards new functions by mutation and selection. So the next time you notice a hummingbird at a flower, see a cat dash after a rodent, or enjoy a slice of pie, take a moment to appreciate not just the diverse tastes that our palates appreciate, but the diverse evolutionary processes that produced our ability to sense them.
Primary literature:
- Baldwin, M. W., Toda, Y., Nakagita, T., O'Connell, M. J., Klasing, K. C., Misaka, T., ...Liberles, S. D. (2014). Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor. Science. 345: 929-933. Read it »
- Nei, M., Niimura, Y., and Nozawa, M. (2008). The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nature Reviews Genetics. 9: 951-963. Read it »
- Shi, P., and Zhang, J. (2006). Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Molecular Biology and Evolution. 23: 292-300. Read it »
News articles:
- A blog post on research about hummingbirds' sense of taste from National Geographic
- An article about the evolution of bitter taste receptors in humans from Science Daily
Understanding Evolution resources:
-
- In your own words, explain how a change in our DNA can affect what we taste. Be sure to include proteins in your explanation
- Review the information about evolutionary change on this page. Which of the four types of change are exemplified in the article above about the evolution of taste? Explain your answer.
- The article above outlines how duplication and divergence produced a wide variety of bitter-sensing genes. Research and describe another example of evolution via duplication and divergence.
- Describe what T1R2 does and how the gene differs among humans, cats, and chickens.
- In hummingbirds, the T1R1 and T1R3 genes evolved in ways that allowed them to sense sweet flavors instead of umami ones. Research and describe another example of a gene evolving a new function.
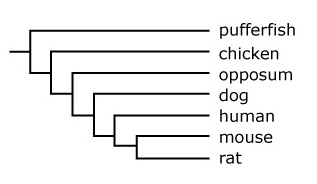
- Advanced: Compare the gene tree in the article above to the evolutionary tree below, which shows the relationships among the lineages of the fish, birds, marsupials, dogs, humans, mice, and rats. How and why are the gene tree and the phylogenetic tree below similar and different?
- Teach about the gain and loss of genes: This 13-minute film for grades 6-16 describes how scientists have pieced together the Antartic icefish's evolutionary history, which involved both the gain and loss of genes leading to key adaptations.
- Teach about duplication and divergence: This short slide set for undergraduates explains uniformity and variation in the process of photosynthesis across all life using evolutionary history. Save the slide set to your computer to view the explanation and notes that go along with each slide.
- Teach about the evolution of another sense: This case study for undergraduates examines the evolution of trichromatic vision in old world monkeys.
- Baldwin, M. W., Toda, Y., Nakagita, T., O'Connell, M. J., Klasing, K. C., Misaka, T., ...Liberles, S. D. (2014). Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor. Science. 345: 929-933.
- Blair, J. E. and Blair Hedges, S. (2005). Molecular phylogeny and divergence times of deuterostome animals. Molecular Biology and Evolution. 22: 2275-2284.
- Nei, M., Niimura, Y., and Nozawa, M. (2008). The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nature Reviews Genetics. 9: 951-963.
- Shi, P., and Zhang, J. (2006). Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Molecular Biology and Evolution. 23: 292-300.