For the past month, news of the pandemic coronavirus, known as COVID-19, has rattled people across the U.S. and around the globe. Internationally, the nearly 1.2 million cases have resulted in more than 60,000 deaths. Businesses have shuttered, jobs have disappeared, large swaths of the population have been asked to shelter in their homes, and many others have been left homeless, hungry, and scared. Our last Evo in the news story explained how evolutionary theory helps answer important questions about the origin of the pandemic. This month we’ll dig even deeper into the virus’s evolutionary history and how those deep relationships relate to prospects for a vaccine.
Where's the evolution?
Structurally, viruses are little more than sacks of genetic material. They consist of DNA or RNA surrounded by a protective shell of proteins, and sometimes a layer of lipids as well. They are so small that they can’t be seen with regular microscopes; scientists must use techniques like electron microscopy to visualize them. Most importantly, viruses can’t reproduce by themselves; they invade the living cells of another organism and use the cellular machinery of that other species to copy themselves and their genetic material.
Viruses seem to occupy the gray zone between biology and chemistry. Not surprisingly, some scientists don’t even consider viruses to be living things. However, being alive is not a prerequisite for experiencing evolution. Any group that varies from one individual to the next and passes on traits to descendents through reproduction (i.e., displays inheritance) can evolve. Computer programs evolve if they are designed to copy themselves with minor variations. Human languages evolve as they change slightly with culture and are passed down to children. Self-duplicating molecules in a test tube evolve as they make copying errors and introduce variation to their lineages. And so, of course, viruses — whose genetic material is imperfectly copied by the machinery of their host cells — also evolve.
When evolving entities like viruses pass heritable information from parent to offspring, we can depict their evolution in a tree shape, called a phylogeny. As variations are introduced and passed to offspring, new branches of the tree form. We can usually reconstruct the evolutionary history of a group by collecting information about the distribution of traits (genetic sequences, anatomical characteristics, behaviors, etc.) among lineages in that group. However, when new variations (e.g., mutations) are introduced rapid fire, it is harder to reconstruct ancient evolutionary relationships. This is because those variations are evidence, and when a lot of them occur, new changes are likely to overwrite older ones, destroying evidence of older relationships. Since viruses mutate quickly, it is very difficult to reconstruct their deep evolutionary history. Nevertheless, scientists have made a lot of progress on this challenge, especially by including information about the structure of viral proteins (and not just genetic sequences) in their analyses.
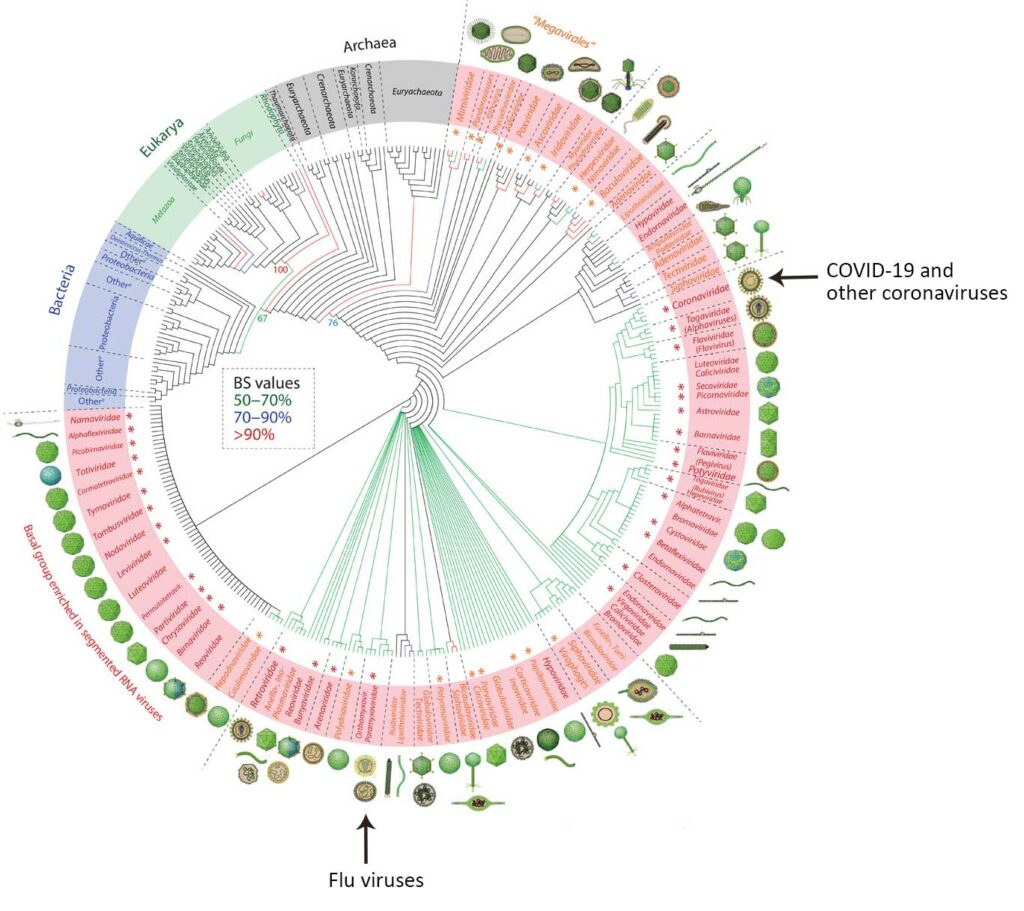
In the family tree of viruses (see below), COVID-19 fits on the branch occupied by other coronaviruses. Notice that coronaviruses are only distantly related to flu viruses, though the range of symptoms they cause is similar. Because they are not closely related, we wouldn’t expect the proteins on the surface of COVID-19 to have much similarity with those of flu virus particles. Vaccines protect us from disease by teaching our immune systems to recognize and fight pathogens based on their surface proteins. In practical terms, this means that flu vaccines are designed to target different surface proteins and simply won’t work against the new pandemic coronavirus, however similar the symptoms might seem. Nevertheless, keeping up to date on your flu shot is especially important during the current pandemic because people who get sick with the flu add even more burden to our stressed healthcare systems.
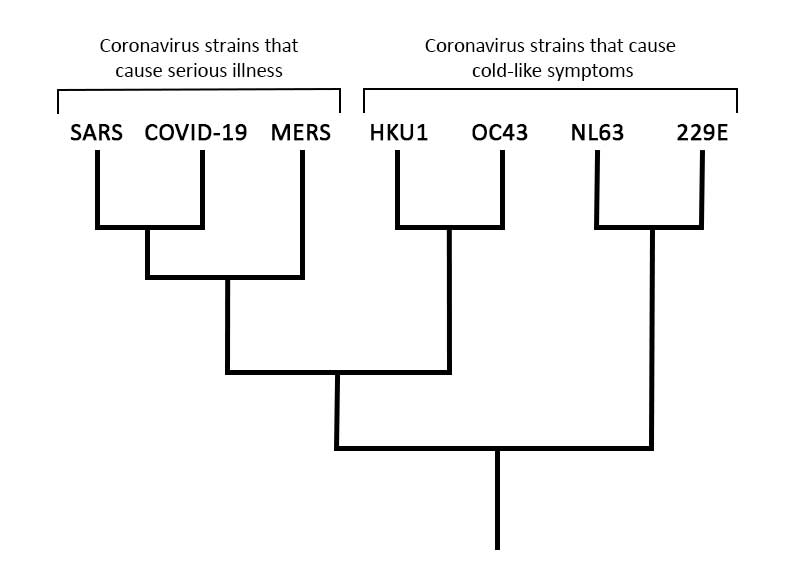
However, digging deeper into the evolutionary relationships among coronaviruses (see tree below) does reveal some cause for hope. Coronaviruses that infect humans cause a range of effects. Some, like the HKU1 strain, cause no more than a cold. Because the effects of these strains are so mild, there has been no motivation to develop vaccines against them. But others, like MERS and the SARS virus that emerged in the early 2000s, are severe diseases that kill a higher percentage of those infected than does COVID-19. SARS was stamped out a few years after it started infecting humans, but researchers did make some progress towards developing a vaccine while it was still a threat. And scientists have continued to work on a MERS vaccine, reaching hopeful milestones, though not a functional vaccine. This work on close relatives of COVID-19 may inform some efforts to develop a COVID-19 vaccine.
A critical factor in creating a vaccine to fight a new pathogen is rate of evolutionary change. Pathogens that evolve slowly (e.g., because they have a low mutation rate) remain quite similar from one infection to the next, presenting a steady target for a vaccine to aim at. However, pathogens that evolve quickly can diversify into a multitude of lineages quickly, forming a bushy evolutionary tree. This makes it difficult to design a single vaccine that can recognize and fight all the different branches of the pathogen’s diverse phylogeny. HIV, for example, has the highest mutation rate of any biological lineage science has ever studied and evolves at lightning speed. For this and other reasons, we still don’t have an HIV vaccine despite more than 35 years of work. Luckily, coronaviruses have a more moderate rate of evolution. While developing an effective vaccine for COVID-19 will still be a challenge, researchers are hopeful.
Understanding COVID-19 as one strain among a multitude of closely related coronaviruses can also give us an idea of how long immunity, once acquired through a vaccine or infection, might last. Coronaviruses that cause cold-like symptoms in humans trigger only short-lived immunity: you can be reinfected by the same virus in less than a year. However, for SARS and MERS — strains more closely related to COVID-19 and strains that cause serious illness, as COVID-19 does — immunity seems to last multiple years (though not a lifetime). While we must wait for more data to be sure, a good starting hypothesis is that immunity to COVID-19 would last several years and that we might need to be revaccinated after that.
The new coronavirus is undoubtedly frightening and dangerous — but it is not unknown or unknowable. Scientific research has built a robust knowledge of viruses, with different facets of their workings illuminated by epidemiology, microbiology, genetics, and of course, evolution. Scientists around the world are bringing all this knowledge to bear as they unite in an effort to develop strategies for infection prevention, treatments, and vaccines.
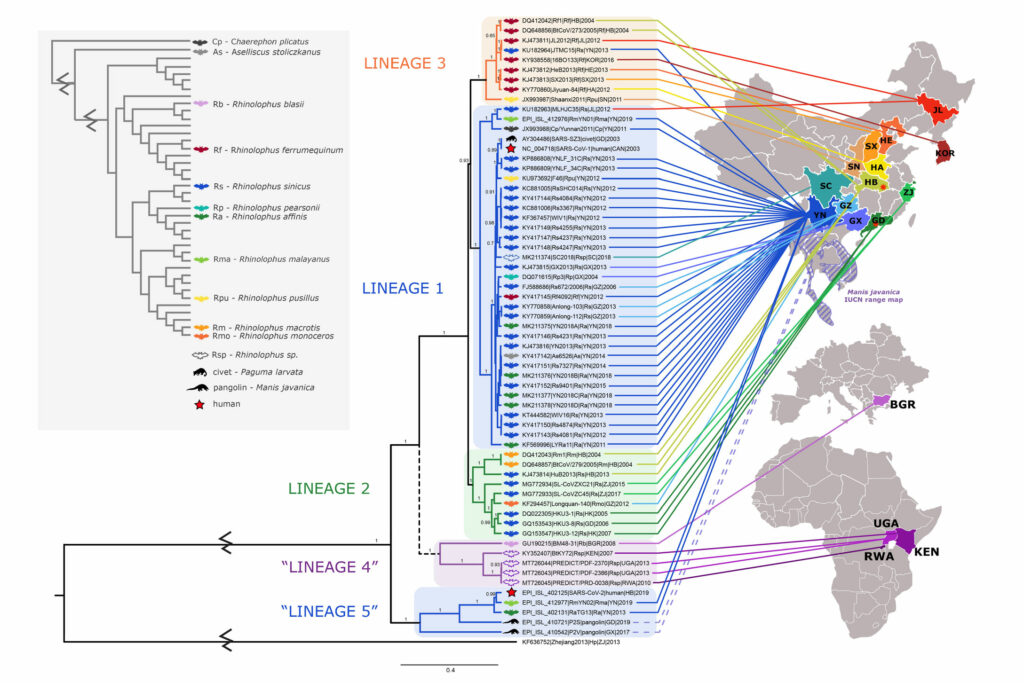
Back in early 2020, when the world was still getting familiar with words like “coronavirus” and “pandemic,” Evo in the News outlined the evolutionary history of SARS-CoV-2, the virus that causes COVID-19. We examined the big picture to see how coronaviruses relate to all life and zoomed in to see how different coronavirus lineages relate to one another. Since then, scientists have continued to investigate the evolution of this pathogen. This past February, a team of biologists announced the results of their deep dive into the closest evolutionary relationships of SARS-CoV-2. This tree summarizes some of the key results. You can see five major clades of coronaviruses (Lineages 1-5 on the diagram), each mainly restricted to a particular area (as shown by the colored lines clustering within regions of Asia and Africa). The diagram also uses icons to show the host animal for each virus (mostly horseshoe bats), with the upper red star representing the virus that caused the previous SARS outbreak and the lower red star representing SARS-CoV-2. The new study suggests that the ability to enter human cells using the ACE2 receptor (a key feature of the viruses causing both SARS outbreaks) has been passed back and forth among coronaviruses via recombination. This highlights the worrisome possibility that the ability to infect humans could arise in other viral lineages relatively easily through recombination. However, monitoring viruses in regions of Asia and Africa where human-infecting coronaviruses have emerged in the past could give us a chance of heading off the next pandemic before it starts.
Primary literature:
- Coronoviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 5: 536-544. Read it »
- Nasir, A., and Caetano-Anollés, G. (2015). A phylogenomic data-driven exploration of viral origins and evolution. Science Advances. 1: e1500527. Read it »
News articles:
- An article and radio program about the progression of the COVID-19 pandemic and vaccine development, from NPR
- An overview of COVID-19 vaccine development efforts from Science Magazine
Understanding Evolution resources:
- Do some research online and explain what characteristics of viruses are consistent with traits of living things, and what characteristics are more consistent with nonliving things.
- COVID-19 infections have many symptoms in common with the flu. Does this mean that the flu vaccine can help fight the new coronavirus? Why or why not?
- Based on the phylogeny in the article above, what other viruses is COVID-19 closely related to?
- Imagine two viruses that are similar, but one has a higher mutation rate than the other due a small difference in the proteins that help copy the each virus’ genetic material. How might this difference affect efforts to develop vaccines against each virus? Explain your reasoning.
- Advanced: Viruses experience horizontal transfer, which means they can pass genetic material directly from one individual to another individual, not just parent-to-offspring during reproduction. How might you represent this process graphically? Would the resulting diagram form a tree-like shape or something else? Explain how one could depict evolutionary relationships among entities that experience both inheritance from parent to offspring and horizontal transfer.
- Teach about emerging infectious disease: In this case study for undergraduates, students explore changes in SIV (Simian Immunodeficiency Virus, thought to be a precursor to HIV) in different chimpanzee populations and how researchers use this information to test hypotheses about the origins of HIV.
- Teach about evolutionary trees: This lab for grades 9-12 has two main parts. In the first, students build phylogenetic trees themed around the evidence of evolution, including fossils, biogeography, and similarities in DNA. In the second, students explore an interactive tree of life and trace the shared ancestry of numerous species.
- Teach about evolution and medicine: In this activity for undergraduates, students use what they know about evolution and medicine to review an article written for a school publication. The task is to identify errors, explain the incorrect statements, and correct the information. They then explain the process of natural selection by creating a labeled illustration using one of the examples from an earlier lesson.
- Cohen, J. (Mar. 31, 2020). With record-setting speed, vaccinemakers take their first shots at the new coronavirus. Science. Retrieved Apr. 4, 2020 from https://www.sciencemag.org/news/2020/03/record-setting-speed-vaccine-makers-take-their-first-shots-new-coronavirus
- Coronoviridae Study Group of the International Committee on Taxonomy of Viruses, The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. (2020). Nature Microbiology. 5: 536-544.
- Cuevas, J. M., Geller, R., Garijo, R., López-Aldeguer, J., and Sanjuán, R. (2015). Extremely high mutation rate of HIV-1 in vivo. PLoS Biology. 13: e1002251.
- Dong, E., Du, H., and Gardner, L. (2020). An interactive web-based dashboard to track COVID-19 in real time. The Lancet. DOI: https://doi.org/10.1016/S1473-3099(20)30120-1. Data retrieved Apr. 4, 2020 from https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6
- Nasir, A., and Caetano-Anollés, G. (2015). A phylogenomic data-driven exploration of viral origins and evolution. Science Advances. 1: e1500527.
- Yong, E. (Mar. 25, 2020). How the pandemic will end. The Atlantic. Retrieved Apr. 4, 2020 from https://www.theatlantic.com/health/archive/2020/03/how-will-coronavirus-end/608719/
