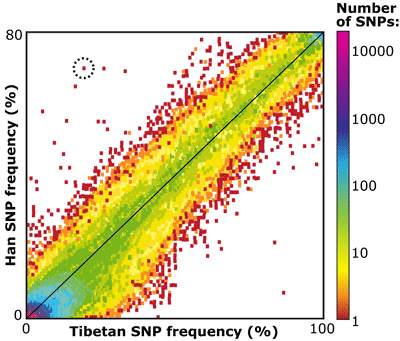
The Chinese team sequenced exons from 50 ethnic Tibetans and 40 Han Chinese, and Emilia and her colleagues got their first look at the alleles present in each. The results are shown below. The x-axis of the graph represents allele frequencies among the Tibetans (ranging from zero to 100 percent) and the y-axis represents the same for the Han (ranging from zero to 80 percent). Points on the graph represent single nucleotide polymorphisms (SNPs) — regions of the genome in which some chromosomes in the population have one nucleotide (e.g., A, adenine) and other chromosomes in the population have a different one (e.g., G, guanine). The colors on the graph correspond to the number of single nucleotide polymorphisms with those frequencies in each population. For example, the point that is circled on the graph represents one single nucleotide polymorphism (we know this because the point is colored red), which is found at a frequency of 20% among Tibetans (we know this from the position along the x-axis) and a frequency of 70% among Han (we know this from the position along the y-axis).
Because the two groups are so closely related, and because there is still some migration between them, the researchers were not surprised to find that, for the most part, the same single nucleotide variations were present in both populations and had similar frequencies. The strong, diagonal pattern in the graph shows this relationship. Alleles that have a high frequency in the Tibetans also tend to have a high frequency in the Han, and likewise for alleles with low frequencies.