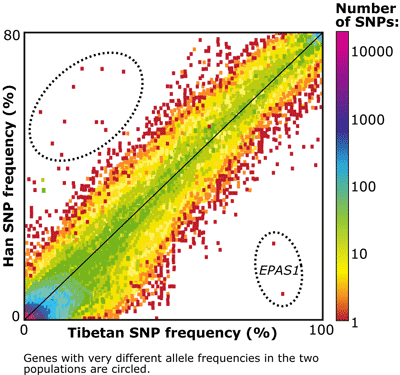
The graph made it clear that some genes stand out from the pattern and have very different allele frequencies in the two populations. These genes have probably undergone selection recently, but Emilia couldn’t be sure which population had experienced selection. The observation of an allele with a high frequency in the Tibetans and low frequency in the Han might have been caused by selection favoring the allele in the Tibetans (and so might be related to adaptation to high altitudes) or by selection acting against the allele the Han (and so have nothing at all to do with adaptation to high altitudes).
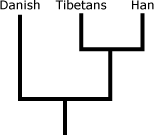 To figure out in which population the selection occurred, the team examined a third, more distantly related European population, the Danish. If, for a particular candidate gene, the Danes and Tibetans have similar frequencies and the Han are outliers, then selection likely occurred in the Han lineage, causing them to differentiate from the other groups. If, however, the Danes and Han have similar allele frequencies and the Tibetans are outliers, then selection likely occurred in the Tibetans and may have been caused by the challenges of living at high altitudes.
To figure out in which population the selection occurred, the team examined a third, more distantly related European population, the Danish. If, for a particular candidate gene, the Danes and Tibetans have similar frequencies and the Han are outliers, then selection likely occurred in the Han lineage, causing them to differentiate from the other groups. If, however, the Danes and Han have similar allele frequencies and the Tibetans are outliers, then selection likely occurred in the Tibetans and may have been caused by the challenges of living at high altitudes.
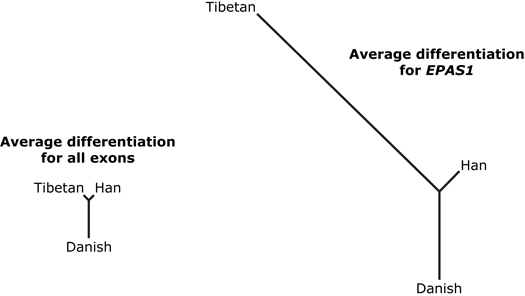
After comparing allele frequencies with the Danish, one gene stood out: EPAS1, short for the mouthful endothelial Per-Arnt-Sim domain protein 1. As you can see on the graph above, two variants of this gene had a frequency of over 80% among Tibetans but less than 30% among Han Chinese. Furthermore, the Han and Danish had similar allele frequencies for this gene, while the Tibetans alone had a high frequency of two unusual alleles — which suggests that selection favored those particular alleles in the Tibetans. In this diagram, the length of each branch is proportional to differentiation in allele frequencies. The left hand diagram shows the average differentiation for all exons, and the right hand diagram shows the differentiation for EPAS1. The extra-long Tibetan branch indicates that, for this gene, allele frequencies changed a lot in the Tibetan population.