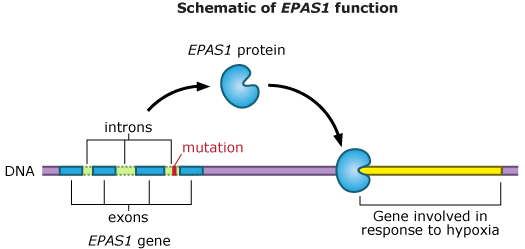
Interestingly, the EPAS1 mutations that are common among Tibetans but rare in the Han are not in the protein-coding regions of the gene, but in an intron, which is spliced out of the gene’s product. Though the team’s initial approach had been to focus on the exons, some portions of introns happened to be included in their sequencing. Emilia and her colleagues were lucky to catch these mutations. Their exon-focused analysis might easily have missed them!
Since their original study was published in 2010, Emilia and her colleagues have continued to investigate EPAS1 — this time focusing on the entire gene, and not just the exons. They’ve discovered that the Tibetan version of the gene differs from the Han’s not by just two base pairs, but by many base pairs over a large region of the intron. In the team’s original study, most of this intron was missed.
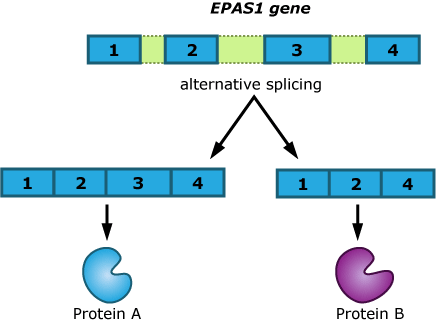
It might seem strange that changes in an intron — a DNA sequence that never makes it into the final product of the gene — could affect the way a gene functions. The team is still trying to figure out exactly how EPAS1‘s introns work. Says Emilia, “One hypothesis is that it is related to alternative splicing.” Perhaps the Tibetan intron directs an alternative splicing pattern that puts the exons together in a slightly different way from how they are put together in the Han, resulting in a different protein. The altered protein could affect when and how EPAS1 turns different genes off and on.
Check out the Human Genome Browser for yourself.