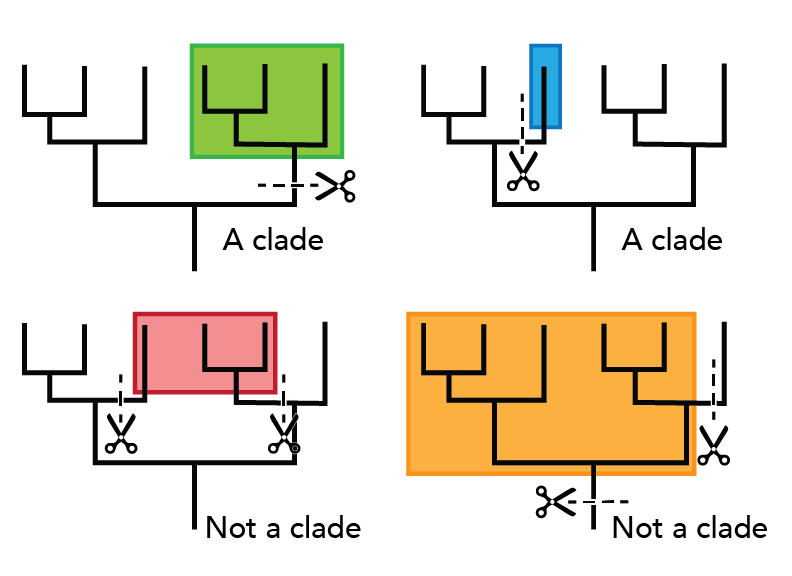
A clade (also known as a monophyletic group) is a group of organisms that includes a single ancestor and all of its descendents. Clades represent unbroken lines of evolutionary descent. It’s easy to identify a clade using a phylogenetic tree. Just imagine clipping any single branch off the tree. All the lineages on that branch form a clade. If you have to make more than one cut to separate a group of organisms from the rest of the tree, that group does not form a clade. Such non-clade groups are called either polyphyletic or paraphyletic groups depending on which taxa they include.
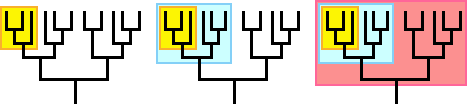
Test your understanding of clades with the tree shown here. Is the group highlighted in blue a clade? Is the group highlighted in yellow a clade? Click the button to see the answer.
Notice that clades are nested within one another. Smaller clades are encompassed by larger ones. For example, the human species forms a clade. It is a single branch within the larger clade of the hominin lineage, which is a single branch within the larger clade of the primate lineage, which is a single branch within the larger clade of the mammalian lineage, and so on, back to the most encompassing clade of all: the entire tree of life.
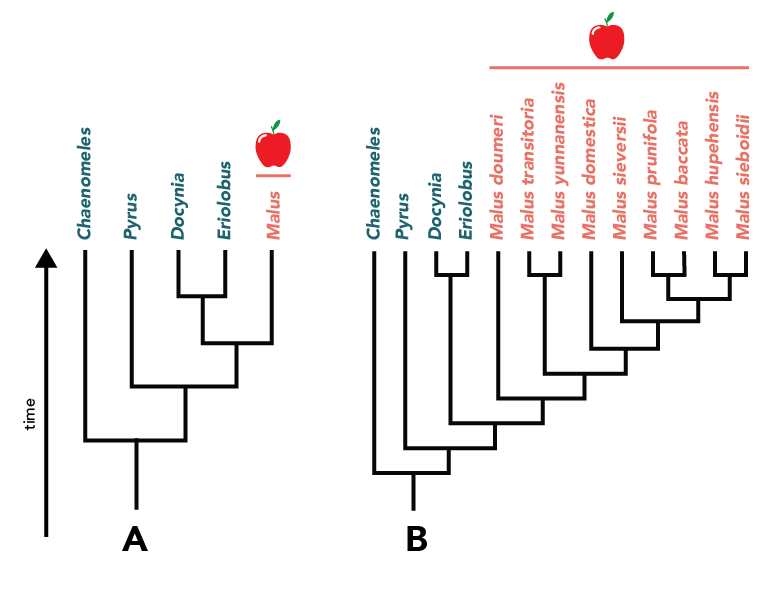
Reader beware! Sometimes trees that represent the same relationships look different from one another. That’s because it’s common practice to prune or collapse clades in order to emphasize certain aspects of a tree. For example, trees A and B below show the same relationships, but in tree A the apple genus is collapsed and in tree B it is expanded to show species relationships.
Similarly, trees C and D are consistent with one another; however, in tree C extinct groups are included, and these groups are pruned in tree D.
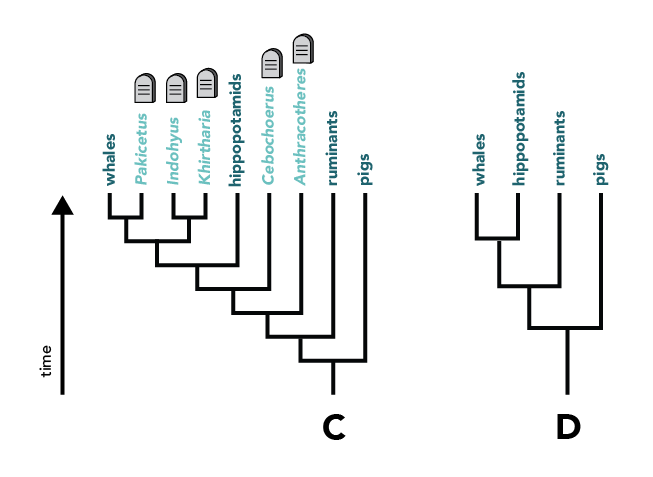
Trees adapted from Lou, G., Wang, S., Zhang, B., Cheng, Y., Wang, H., 2020. The complete chloroplast genome sequence of Malus sieboldii (Rosaceae) and its phylogenetic analysis. Mitochondrial DNA. Part B 5, 2170–2171. https://doi.org/10.1080/23802359.2020.1768940; Xiang, Y., Huang, C.-H., Hu, Y., Wen, J., Li, S., Yi, T., Chen, H., Xiang, J., Ma, H., 2016. Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol Biol Evol. msw242. https://doi.org/10.1093/molbev/msw242; Gatesy, J., Geisler, J.H., Chang, J., Buell, C., Berta, A., Meredith, R.W., Springer, M.S., McGowen, M.R., 2012. A phylogenetic blueprint for a modern whale. Molecular Phylogenetics and Evolution. 66, 479–506. https://doi.org/10.1016/j.ympev.2012.10.012