Phylogenies can also help us figure out which species are likely to have inherited a particular trait — such as the ability to produce a medically useful substance. Drug researchers are always on the lookout for compounds that are biologically active — those that have some (hopefully beneficial!) effect on the human body. Usually, that means beginning their hunt in the biological world. After all, evolution has had 3.5 billion years to produce a wide range of chemicals. The basic compounds for treating high blood pressure, pain, and many other ailments can be found in nature; the main problem is sorting through the enormous number of species that might harbor just the right substance. Phylogenetics offers us a way to organize species and focus on the ones that are likely to produce useful compounds. For example, when scientists discovered that paclitaxel (i.e., Taxol) could be used to treat some types of cancer, they were hopeful that it could save lives. However, the chemical had to be harvested from the bark of the Pacific Yew tree (Taxus brevifolia) — a process which kills the tree and, if scaled up, threatened to drive the species extinct. The search for a route around this bottleneck included screening species occupying branches of the tree of life adjacent to that of T. brevifolia, which, biologists reasoned, might have inherited the ability to produce the compound from the same ancestor that T. brevifolia did. Happily, they discovered that the needles of the abundant European Yew (T. baccata) produced a related compound that could be used to synthesize paclitaxel.6 Still later, it was discovered that paclitaxel was actually produced by a fungus that lived symbiotically with the tree.7 Now, biologists are studying microbial plant symbionts hunting for new drugs, as well as using phylogenies to focus their attention on plant species with potential medical importance.8
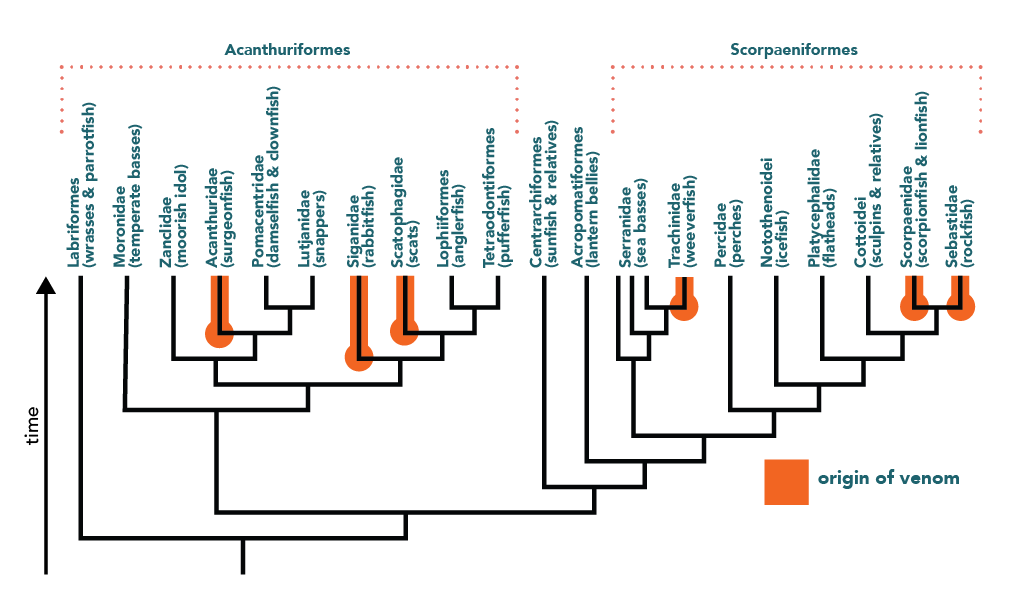
Other particularly fruitful branches of the tree of life for drug discovery are those belonging to venom-producing animals. Venoms may seem like the antithesis of healing substances, but in fact, have yielded several important drugs (e.g., ACE inhibitors, a common high blood pressure medication). Because we know so little about the natural history of most animals, many venoms have yet to be discovered. To find new venoms then, biologists turn to phylogenetics. Venom (like carrying paclitaxel-producing fungi) is a heritable trait that is often shared among closely related species. Biologists can reconstruct the phylogenetic tree of a group of species, map onto the tree those that are known to be venomous, and use the pattern of evolution to identify other species on the tree that are likely to have inherited this trait. When biologists used this approach to study fish, they discovered that more than 1200 species of fish not known to be venomous probably are!9 This represents a treasure trove of potentially useful chemicals for drug developers. Similar phylogenetic approaches may help us develop drugs from snake, lizard, and snail venom.10
Feeling lost? Review tree basics with the primer.
6 Flam, F. 1994. Race to synthesize taxol ends in a tie. Science 263:911.
7 Saslis-Lagoudakis, C.H., V. Savolainen, E.M. Williamson, F. Forest, S.J. Wagstaff, S.R. Baral, … and J.A. Hawkins. 2012. Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proceedings of the National Academy of Sciences USA. DOI: 10.1073/pnas.1202242109
Stierle, A., G. Strobel, and D. Stierle. 1993. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 260:214-216.
8 Strobel, G., and B. Daisy. 2003. Bioprospecting for microbial endophytes and their natural products. Microbiology and Molecular Biology Reviews 67:491-502.
9 Smith, W.L., and W.C. Wheeler. 2006. Venom evolution widespread in fishes: A phylogenetic road map for the bioprospecting of piscine venoms. Journal of Heredity 97:206-217.
10 Espiritu, D.J.D., M. Watkins, V. Dia-Monje, G.E. Cartier, L.J. Cruz, and B.M. Olivera. 2001. Venomous cone snails: Molecular phylogeny and the generation of toxin diversity. Toxicon 39:1899-1916.
Schroeder, C.I., and D.J. Craik. 2012. Therapeutic potential of conopeptides. Future Medicinal Chemistry 4:1243-1255.
Fry, B.G., K. Roelants, K. Winter, W.C. Hodgson, L. Griesman, H. Fai Kwok, … and J.A. Norman. 2010. Novel venom proteins produced by differential domain-expression strategies in beaded lizards and Gila monsters (genus Helodroma). Molecular Biology and Evolution 27:395-407.
Fry, B.G., W. Wüster, R.M. Kini, V. Brusic, A. Khan, D. Venkataraman, and A.P. Rooney. 2003. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. Journal of Molecular Evolution 57:110-129.
Holford, M., N. Puillandre, M.V. Modica, M. Watkins, R. Collin, E. Bergminham, and B.M. Olivera. 2009. Correlating molecular phylogeny with venom apparatus occurrence in Panamic auger snails (Terebridae). PLoS One 4:e7667.
Tree adapted from Smith, W.L., Stern, J.H., Girard, M.G., Davis, M.P., 2016. Evolution of Venomous Cartilaginous and Ray-Finned Fishes. Integr. Comp. Biol. 56, 950–961. https://doi.org/10.1093/icb/icw070; Smith, W.L., Wheeler, W.C., 2006. Venom Evolution Widespread in Fishes: A Phylogenetic Road Map for the Bioprospecting of Piscine Venoms. Journal of Heredity 97, 206–217. https://doi.org/10.1093/jhered/esj034; Betancur, R., Wiley, E.O., Arratia, G., Acero, A., Bailly, N., Miya, M., Lecointre, G., Ortí, G., 2017. Phylogenetic classification of bony fishes. BMC Evol Biol 17, 162. https://doi.org/10.1186/s12862-017-0958-3
