At the most basic level, evolutionary trees tell us which groups of organisms (i.e., clades) are closely related and which are more distant cousins. This can be useful in a couple of ways. First, if a particular organism is unfamiliar, placing it on an evolutionary tree can reveal its origins. This might seem like a no brainer (after all, if the organism is a dog, it must be closely related to other dogs); however, in practice it is often not so simple. Viruses and bacteria often can’t be identified based on their appearances, but can be placed on a phylogenetic tree based on their genetic sequences. For example, the origins of HIV were a mystery to medical researchers until samples of the HIV virus and many others were sequenced and used to construct a phylogenetic tree. The tree made it obvious that HIV evolved from the Simian Immunodeficiency Virus (SIV) and that, on a few different occasions, SIV had jumped from another primate to a human host and, over many generations, was selected for the traits that we now associate with HIV. HIV evolved not just once, but several different times!1 Clearly, when humans come into close contact with our nearest relatives, the risk of passing infectious diseases to one another is relatively high.
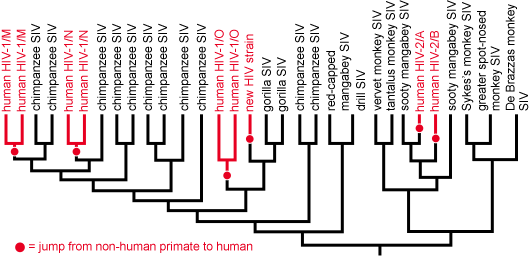
Understanding the relationships among groups of organisms has been useful for much more than just identifying the origins of HIV. The same basic approach has helped uncover the roots of SARS (from bats), malaria (from primates), and other diseases. Understanding how infectious diseases got their starts in humans may help us prevent other animal-borne diseases from starting to infect humans. It also allows researchers to study the disease in other contexts — potentially resulting in new treatments. The same approach has been useful in non-medical contexts as well. For example, whale species being illegally sold in Japanese meat markets and alligator being sold as turtle meat in the southern U.S. have all been identified on the basis of phylogenetic trees linking suspect meat samples to known species.2
Feeling lost? Review tree basics with the primer.
1 Keele, B.F., F. Van Heuverswyn, Y. Li, E. Bailes, J. Takehisa, M.L. Santiago, … and B.H. Hahn. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Nature 313:523-526.
2 C.S. Baker, G.M. Lento, F. Cipriano, M.L. Dalebout, and S.R. Palumbi. (2000). Scientific whaling: Source of illegal products for market? Science 290:1695.
Baker, C.S., and S.R. Palumbi. 1996. Population structure, molecular systematics and forensic identification of whales and dolphins. In J.C. Avise and J.L. Hamrick (eds.), Conservation Genetics: Case Histories from Nature. Chapman and Hall.
Palumbi, S.R., and F. Cipriano. 1998. Species identification using genetic tools: The value of nuclear and mitochondrial gene sequences in whale conservation. Journal of Heredity 89:459-464.
Roman, J., and B.W. Bowen. 2000. The mock turtle syndrome: Genetic identification of turtle meat purchased in the southeast United States. Animal Conservation 3:61-65.
