If a biologist wants to build an evolutionary tree for a group (say, pine trees or New World monkeys), the first thing they will do is gather evidence (in the form of DNA, morphology, physiology, and/or behavioral traits) about the individual species. That evidence is expressed in the form of characters — heritable features that can be compared across organisms. For most sorts of traits, it’s obvious how they should be compared — the femur length in one species should be compared to the femur length in another, the extent of parental care in one species should be compared to the extent of parental care in another, the number of vascular bundles in pine needles of one species should be compared to the number in another’s needles, etc. — but for DNA sequences, this is less obvious. Because DNA is made up of a sequence of only four possible bases (A, T, C, or G), it can be hard to tell which positions in the sequences of the different species should be compared to one another. To solve this problem, biologists align the sequences first, inserting blank spaces in order to line the sequences up based on regions of similarity. These blank spaces represent mutations that added or subtracted bits of DNA at some point in evolutionary history. Sequence alignment allows biologists to determine which positions are comparable.
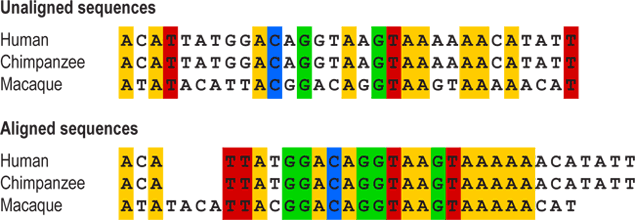
So to build a phylogenetic tree of pines, a biologist might start by sequencing the same set of genes in each pine species and aligning them. In that case, each position in each gene would represent a different character. Alternatively (or in addition to the DNA sequences), the biologist might collect information about the same set of physical traits for each species: whether it has thick or thin bark, whether its seeds have “wings” for wind dispersal, etc. In that case, each individual trait represents a character. Trees based on many characters are more likely to be accurate than trees based on just a few, so taking the time to gather enough evidence and select the right characters is a key step of tree building.
Feeling lost? Review tree basics with the primer.
3 Sequence data from Knowles, D.G., and A. McLysaght. 2009. Recent de novo origin of human protein-coding genes. Genome Research 19:1752-1759.
