Biologists use several different methods for reconstructing evolutionary trees. Parsimony analysis, discussed previously, is easy to explain and is commonly used; however, for complex molecular data sets, evolutionary biologists often turn to two other methods: Maximum Likelihood and Bayesian analyses. Based on probability theory, these are mathematical methods for building trees that can be used when something is known about the process of evolution underlying the data set — i.e., when we have a “model” of how evolution works. For example, because of the structure of DNA, genetic mutations that change an A base to a G base are much more common than mutations that change an A base to a T or C base. Maximum Likelihood and Bayesian analyses can take this into account, generating trees that are more likely to be accurate based on biochemical information about which bases are likely to mutate into one another. Though the method used to reconstruct a tree (parsimony, Likelihood, or Bayesian) may affect the shape of the best-supported tree to some degree, different methods usually agree on the basic structure of the tree and additional data can then be used to sort out discrepancies.
Bayesian analyses are particularly useful because instead of producing a single tree that best fits the data, they will produce a collection of trees that cover a range of likely histories. This is particularly useful to biologists, because it can show them multiple alternative evolutionary histories and suggest when rare events – like hybridization – might have happened. These can help biologists understand why certain tree shapes might be more likely than others, and automatically tells them how much trust to put in their best-supported tree.
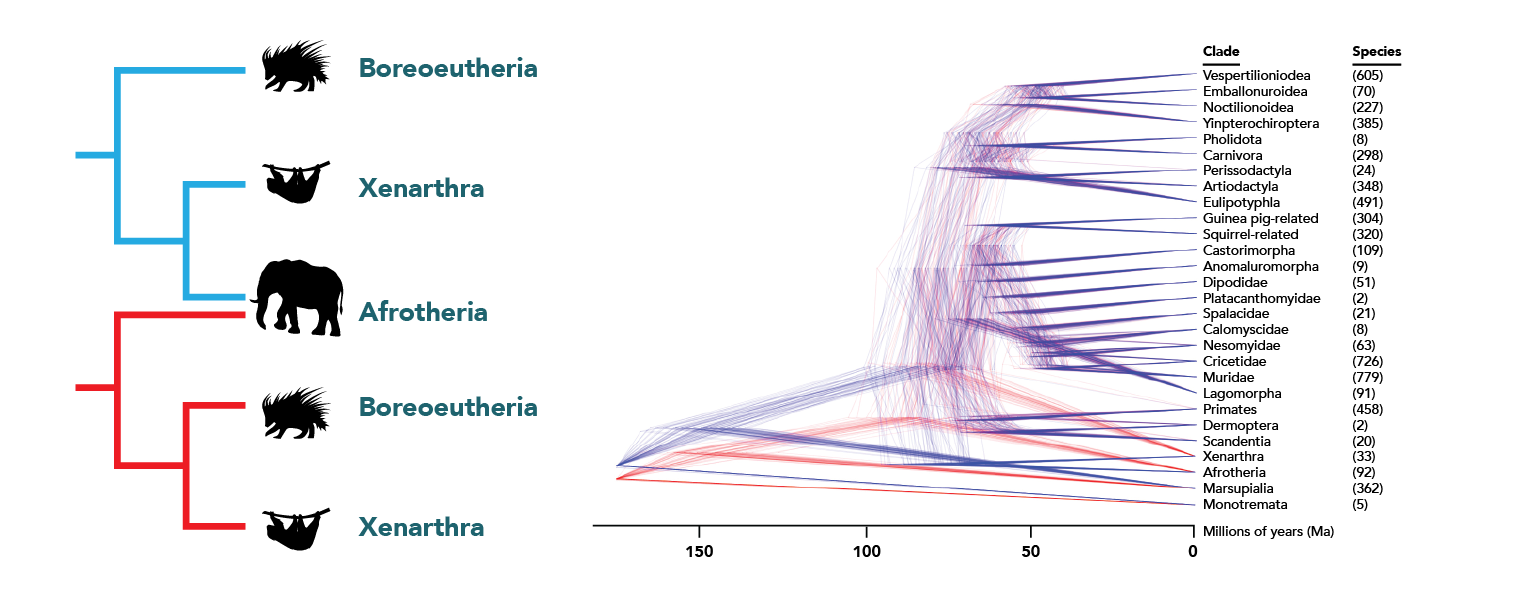
For example, the diagram below is based on a Bayesian analysis and shows several hundred possible versions of the tree of all living mammals as a cloud of trees. The trees on the left show the two possibilities for the shape of the placental mammals and their relative probabilities of being true. In trees colored blue, the clade Xenarthra (sloths, armadillos, and anteaters) is most closely related to the clade Afrotheria (elephants, manatees, and their relatives). In trees colored red, Xenarthra is closest to the Boreoeutheria, a clade that unites all of the rest of the placental mammals (represented by the porcupine). There are only slightly more blue trees than red, meaning that it is slightly more likely that Afrotheria and Xenarthra form a clade together.
Feeling lost? Review tree basics with the primer.
