Research shows that some students, teachers, and members of the general public have intuitive ideas about evolutionary trees that make them likely to misinterpret trees in predictable ways. The following tips, gleaned from the research literature, can help you design evolutionary trees that are less likely to be misinterpreted. For more details about tree-related misconceptions, visit our page on that topic.
- If your branching diagram includes named groups or species on internal nodes and branches, ensure that this is scientifically accurate before using the diagram (i.e., make sure that you intend to depict an ancestor-descendant relationship). In many cases, a true evolutionary tree with named taxa only at the tips is the scientifically correct way to convey these relationships. 1, 2, 3
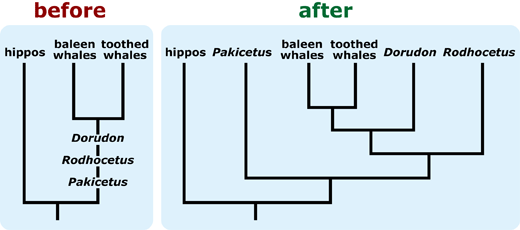
- Draw your tree with horizontal and vertical lines. Tree with diagonal lines are more difficult to interpret. If you need to use a tree with diagonal lines, draw the phylogeny with the outgroup positioned on the right side of the diagram, and consider including shared characters (specifically, synapomorphies) on the phylogeny, as these changes ease interpretation. 4, 5, 6, 7
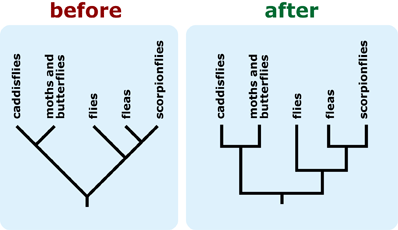
- Rotate branches around nodes to avoid a ladderized tree — one in which the most taxon-rich clades are consistently positioned to the right of less diverse clades on vertically-oriented trees or above less diverse clades on horizontally-oriented trees. This can help discourage intuitive notions of evolutionary progress or advancement. 8, 9
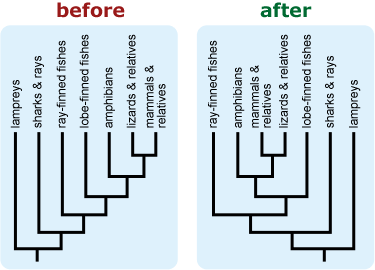
- Rotate branches around nodes to avoid a tree in which groups of interest or those that are commonly perceived as more “advanced” or extreme appear at the right side of a vertically-oriented tree or top of a horizontally-oriented tree. This may be particularly important if your tree includes humans because placing humans in such a “privileged” location tends to trigger intuitive conceptions about progressive evolution. 10, 11
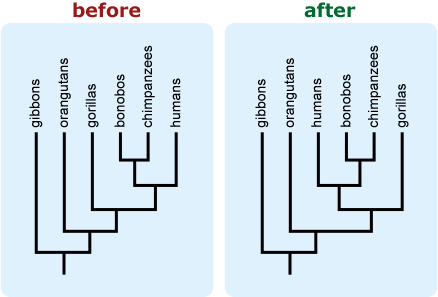
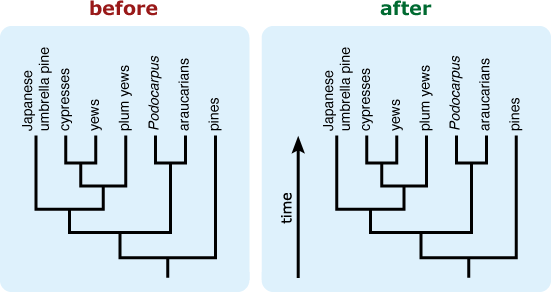
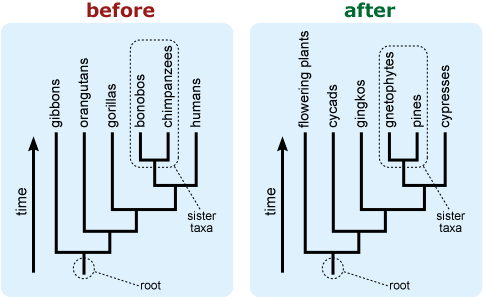
- Include an “arrow of time” on the tree leading from the root toward the tips. This can help combat the common misconception that time is read across the tips of a tree. 9,12,13,14
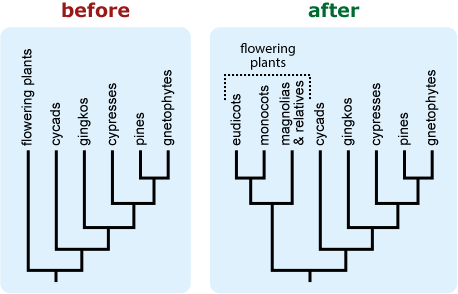
- If a tree is likely to be misinterpreted as a “main line” of evolution with “side tracks,” consider including additional representatives of the “side track” lineages to better reflect their diversity. 15
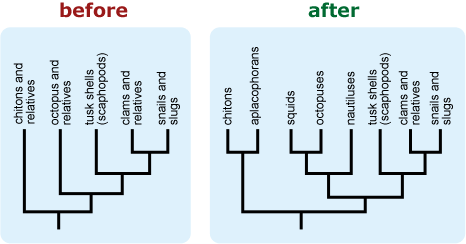
- If a tree is falsely “unbalanced” (i.e., depicts a diverse basal lineage as sparse in comparison to its sister taxon), provide some indication of the true diversity of the basal lineage. This can help combat notions that some extant (i.e., living) lineages are “primitive.” 16
- Be explicit about what species or groups are represented by terminal taxa. The silhouette of a mouse may be intended to represent the clade made up of all mammals, but students may interpret this as representing rodents specifically.
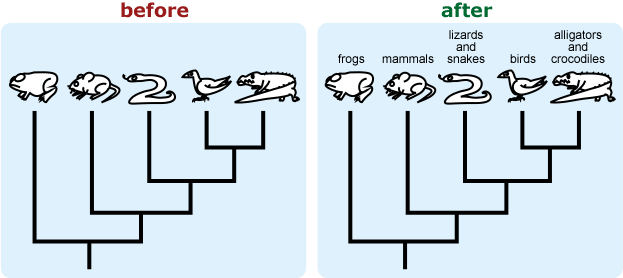
- If you want to emphasize the graphic features of a tree (i.e., focus on tree-reading skills), present a tree with unfamiliar taxa. People tend to ignore the graphic features of a tree when they already have prior knowledge about the taxa in the tree or have intuitive ideas about the relationships among taxa (usually based on superficial similarity); they may then rely on their prior knowledge and misinterpret the tree. 10, 12, 17
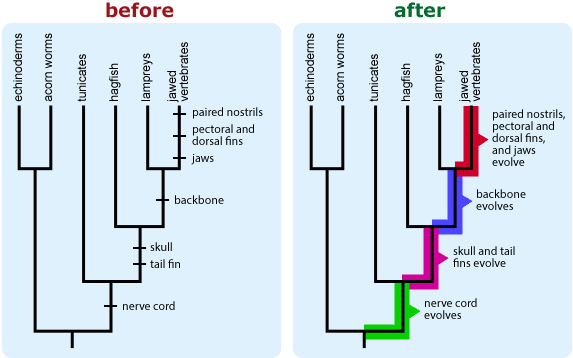
- If your tree depicts character state changes (e.g., the evolution of tail fins) and if more than one evolutionary change maps to a single branch segment, be aware that readers are likely to interpret the order of character state changes shown on that branch as biologically meaningful, even though in most cases, we cannot know the order in which changes occurred. An alternate way of indicating character state changes may help combat this confusion.
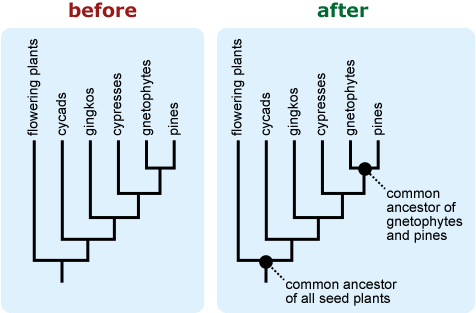
- Labeling nodes or the root of the tree as “shared ancestor” or “common ancestor” may help people correctly interpret patterns of ancestry represented by the phylogeny. 18
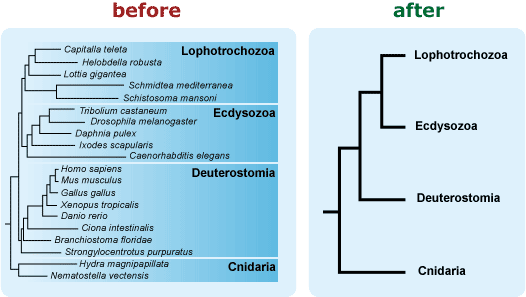
- Avoid including extraneous information on your phylogeny. For example, if branch length is not important to the message you want your tree to convey, use a phylogeny in which branch length doesn’t vary. Readers have enough challenges correctly interpreting the relevant graphical features of a phylogeny, and “extra” features just make the task trickier. 13, 19
Visit our page on tree-related misconceptions.
1 Catley, K.M., L.R. Novick, and C.K. Shade. 2010. Interpreting evolutionary diagams: When topology and process conflict. Journal of Research in Science Teaching 47(7):861-882.
2 Novick, L.R., C.K. Shade, and K.M. Catley. 2011. Linear versus branching depictions of evolutionary history: Implications for diagram design. Topics in Cognitive Science 3:536-559.
3 Bajpai, S., J.G.M. Thewissen,and A. Sahni, A. 2009. The origin and early evolution of whales: macroevolution documented on the Indian Subcontinent. Journal of Bioscience 34:673-686.
4 Novick, L.R., and K.M. Catley. 2007. Understanding phylogenies in biology: The influence of a gestalt perceptual principle. Journal of Experimental Psychology: Applied 13:197-223.
5 Novick, L.R., K.M. Catley, and D.J. Funk. 2010. Characters are key: The effect of synapomorphies on cladogram comprehension. Evolution Education and Outreach. Published online January 19.
6 Novick, L.R., A.T. Stull, and K.M. Catley. 2012. Reading phylogenetic trees: Effects of tree orientation and text processing on comprehension. BioScience 62:757-764.
7 Trautwein, M.D., B.M. Wiegmann, R. Beutel, K.M. Kjer, and D.K. Yeates. 2012. Advances in insect phylogeny at the dawn of the postgenomic era. Annual Review of Entomology 57:449-468.
8 Gregory, T.R. 2008. Understanding evolutionary trees. Evolution and Education Outreach 1:121-137.
9 Meir, E., J. Perry, J.C. Herron, and J. Kingsolver. 2007. College students' misconceptions about evolutionary trees. American Biology Teacher 69:71-76.
10 Diamond, J., and E.M. Evans. 2007. Museums teach evolution. Evolution 61(6):1500-1506.
11 Phillips, B.C., L.R. Novick, and K.M. Catley. 2013. The great chain of being: How diagram design may mislead students to reason teleologically about human evolution. Manuscript in preparation.
12 Dodick, J. 2010. Phylogeny exhibits and understanding geological time. Paper presented at the Understanding the Tree of Life, Carnegie Museum of Natural History, Pittsburgh, PA.
13 Catley, K.M., and L.R. Novick. 2008. Seeing the wood for the trees: An analysis of evolutionary diagrams in biology textbooks. BioScience 58(10):976-987.
14 Ran, J., H. Gao, and X. Wang. 2010. Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Molecular Phylogenetics and Evolution 54:136-149.
15 Kocot, K.M., J.T. Cannon, C. Todt, M.R. Citarella, A.B. Kohn, A. Meyer, … and K.M. Halanych. 2011. Phylogenomics reveals deep molluscan relationships. Nature 477:452-457.
16 Soltis, P.S., and D.E. Soltis. 2013. Angiosperm phylogeny: A framework for studies of genome evolution. In I.J. Leitch et al. (eds.), Plant Genome Diversity Volume 2. Springer-Verlat, Wien.
17 Novick, L R., and K.M. Catley. 2012. When relationships depicted diagrammatically conflict with prior knowledge: An investigation of students' interpretations of evolutionary trees. Invited resubmission under review.
18 Donovan, S., and L. Wilcox. 2004. Tree figures in texts: A framework for unpacking their educational potential. Poster presented at the Society for the Study of Evolution meeting, Fort Collins, CO.
19 Simakov, O., F. Marletaz, S. Cho, E. Edsinger-Gonzales, P. Havlak, U. Hellston, … and D.S. Rokhsar. 2012. Insights into bilaterian evolution from three spiralian genomes. Nature. doi:10.1038/nature11696
